Kashif Rajpoot
Handcrafted Histological Transformer (H2T): Unsupervised Representation of Whole Slide Images
Feb 14, 2022



Abstract:Diagnostic, prognostic and therapeutic decision-making of cancer in pathology clinics can now be carried out based on analysis of multi-gigapixel tissue images, also known as whole-slide images (WSIs). Recently, deep convolutional neural networks (CNNs) have been proposed to derive unsupervised WSI representations; these are attractive as they rely less on expert annotation which is cumbersome. However, a major trade-off is that higher predictive power generally comes at the cost of interpretability, posing a challenge to their clinical use where transparency in decision-making is generally expected. To address this challenge, we present a handcrafted framework based on deep CNN for constructing holistic WSI-level representations. Building on recent findings about the internal working of the Transformer in the domain of natural language processing, we break down its processes and handcraft them into a more transparent framework that we term as the Handcrafted Histological Transformer or H2T. Based on our experiments involving various datasets consisting of a total of 5,306 WSIs, the results demonstrate that H2T based holistic WSI-level representations offer competitive performance compared to recent state-of-the-art methods and can be readily utilized for various downstream analysis tasks. Finally, our results demonstrate that the H2T framework can be up to 14 times faster than the Transformer models.
Towards Launching AI Algorithms for Cellular Pathology into Clinical & Pharmaceutical Orbits
Dec 17, 2021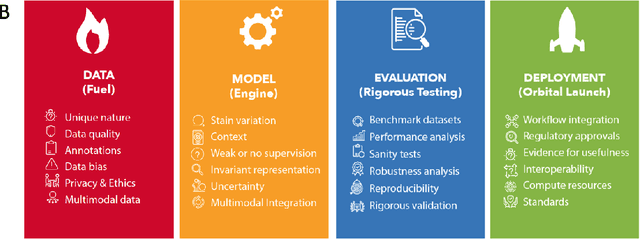

Abstract:Computational Pathology (CPath) is an emerging field concerned with the study of tissue pathology via computational algorithms for the processing and analysis of digitized high-resolution images of tissue slides. Recent deep learning based developments in CPath have successfully leveraged sheer volume of raw pixel data in histology images for predicting target parameters in the domains of diagnostics, prognostics, treatment sensitivity and patient stratification -- heralding the promise of a new data-driven AI era for both histopathology and oncology. With data serving as the fuel and AI as the engine, CPath algorithms are poised to be ready for takeoff and eventual launch into clinical and pharmaceutical orbits. In this paper, we discuss CPath limitations and associated challenges to enable the readers distinguish hope from hype and provide directions for future research to overcome some of the major challenges faced by this budding field to enable its launch into the two orbits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge