Alexander Krull
ShapeEmbed: a self-supervised learning framework for 2D contour quantification
Jul 01, 2025Abstract:The shape of objects is an important source of visual information in a wide range of applications. One of the core challenges of shape quantification is to ensure that the extracted measurements remain invariant to transformations that preserve an object's intrinsic geometry, such as changing its size, orientation, and position in the image. In this work, we introduce ShapeEmbed, a self-supervised representation learning framework designed to encode the contour of objects in 2D images, represented as a Euclidean distance matrix, into a shape descriptor that is invariant to translation, scaling, rotation, reflection, and point indexing. Our approach overcomes the limitations of traditional shape descriptors while improving upon existing state-of-the-art autoencoder-based approaches. We demonstrate that the descriptors learned by our framework outperform their competitors in shape classification tasks on natural and biological images. We envision our approach to be of particular relevance to biological imaging applications.
bit2bit: 1-bit quanta video reconstruction via self-supervised photon prediction
Oct 30, 2024
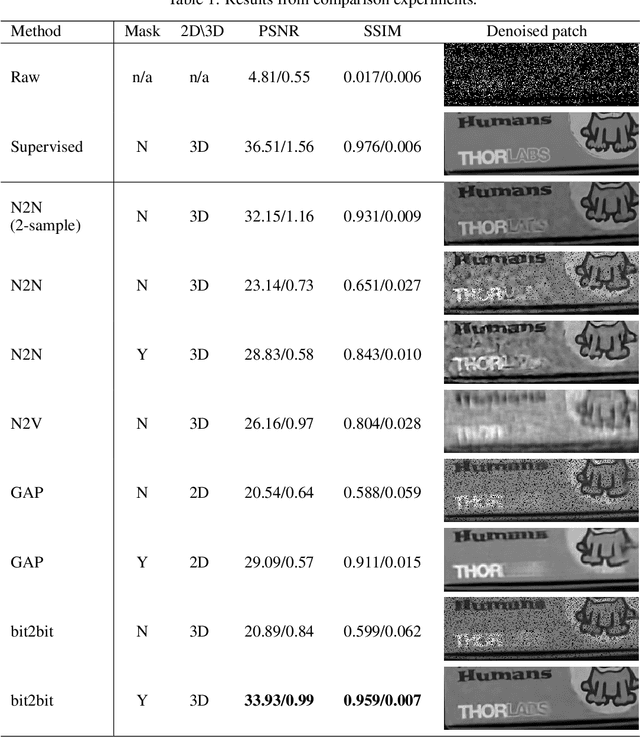


Abstract:Quanta image sensors, such as SPAD arrays, are an emerging sensor technology, producing 1-bit arrays representing photon detection events over exposures as short as a few nanoseconds. In practice, raw data are post-processed using heavy spatiotemporal binning to create more useful and interpretable images at the cost of degrading spatiotemporal resolution. In this work, we propose bit2bit, a new method for reconstructing high-quality image stacks at the original spatiotemporal resolution from sparse binary quanta image data. Inspired by recent work on Poisson denoising, we developed an algorithm that creates a dense image sequence from sparse binary photon data by predicting the photon arrival location probability distribution. However, due to the binary nature of the data, we show that the assumption of a Poisson distribution is inadequate. Instead, we model the process with a Bernoulli lattice process from the truncated Poisson. This leads to the proposal of a novel self-supervised solution based on a masked loss function. We evaluate our method using both simulated and real data. On simulated data from a conventional video, we achieve 34.35 mean PSNR with extremely photon-sparse binary input (<0.06 photons per pixel per frame). We also present a novel dataset containing a wide range of real SPAD high-speed videos under various challenging imaging conditions. The scenes cover strong/weak ambient light, strong motion, ultra-fast events, etc., which will be made available to the community, on which we demonstrate the promise of our approach. Both reconstruction quality and throughput substantially surpass the state-of-the-art methods (e.g., Quanta Burst Photography (QBP)). Our approach significantly enhances the visualization and usability of the data, enabling the application of existing analysis techniques.
WiNet: Wavelet-based Incremental Learning for Efficient Medical Image Registration
Jul 18, 2024Abstract:Deep image registration has demonstrated exceptional accuracy and fast inference. Recent advances have adopted either multiple cascades or pyramid architectures to estimate dense deformation fields in a coarse-to-fine manner. However, due to the cascaded nature and repeated composition/warping operations on feature maps, these methods negatively increase memory usage during training and testing. Moreover, such approaches lack explicit constraints on the learning process of small deformations at different scales, thus lacking explainability. In this study, we introduce a model-driven WiNet that incrementally estimates scale-wise wavelet coefficients for the displacement/velocity field across various scales, utilizing the wavelet coefficients derived from the original input image pair. By exploiting the properties of the wavelet transform, these estimated coefficients facilitate the seamless reconstruction of a full-resolution displacement/velocity field via our devised inverse discrete wavelet transform (IDWT) layer. This approach avoids the complexities of cascading networks or composition operations, making our WiNet an explainable and efficient competitor with other coarse-to-fine methods. Extensive experimental results from two 3D datasets show that our WiNet is accurate and GPU efficient. The code is available at https://github.com/x-xc/WiNet .
Can virtual staining for high-throughput screening generalize?
Jul 09, 2024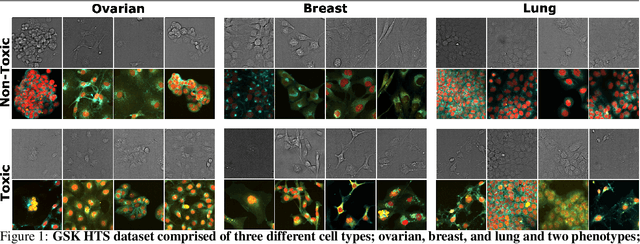
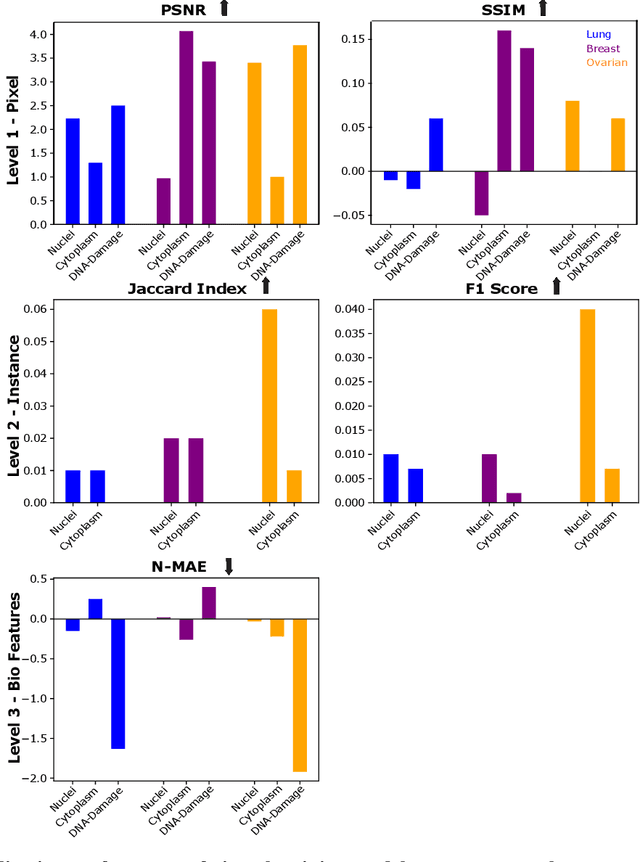
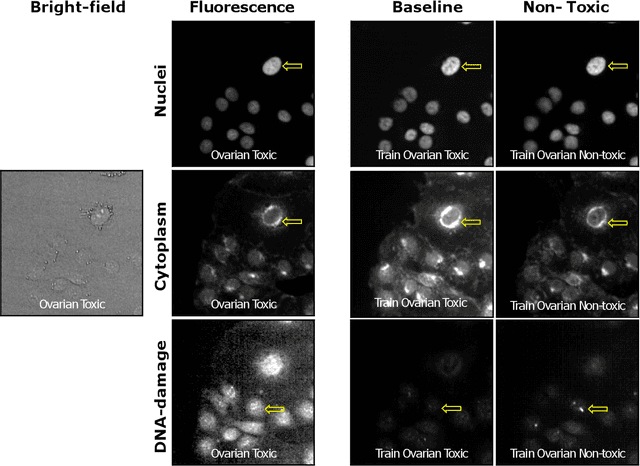
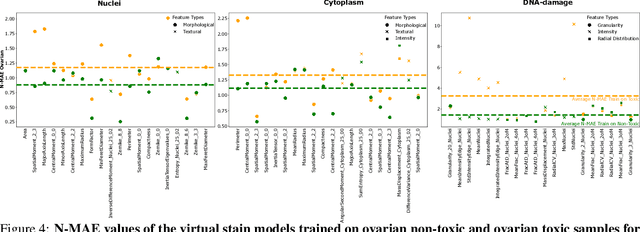
Abstract:The large volume and variety of imaging data from high-throughput screening (HTS) in the pharmaceutical industry present an excellent resource for training virtual staining models. However, the potential of models trained under one set of experimental conditions to generalize to other conditions remains underexplored. This study systematically investigates whether data from three cell types (lung, ovarian, and breast) and two phenotypes (toxic and non-toxic conditions) commonly found in HTS can effectively train virtual staining models to generalize across three typical HTS distribution shifts: unseen phenotypes, unseen cell types, and the combination of both. Utilizing a dataset of 772,416 paired bright-field, cytoplasm, nuclei, and DNA-damage stain images, we evaluate the generalization capabilities of models across pixel-based, instance-wise, and biological-feature-based levels. Our findings indicate that training virtual nuclei and cytoplasm models on non-toxic condition samples not only generalizes to toxic condition samples but leads to improved performance across all evaluation levels compared to training on toxic condition samples. Generalization to unseen cell types shows variability depending on the cell type; models trained on ovarian or lung cell samples often perform well under other conditions, while those trained on breast cell samples consistently show poor generalization. Generalization to unseen cell types and phenotypes shows good generalization across all levels of evaluation compared to addressing unseen cell types alone. This study represents the first large-scale, data-centric analysis of the generalization capability of virtual staining models trained on diverse HTS datasets, providing valuable strategies for experimental training data generation.
Direct Unsupervised Denoising
Oct 27, 2023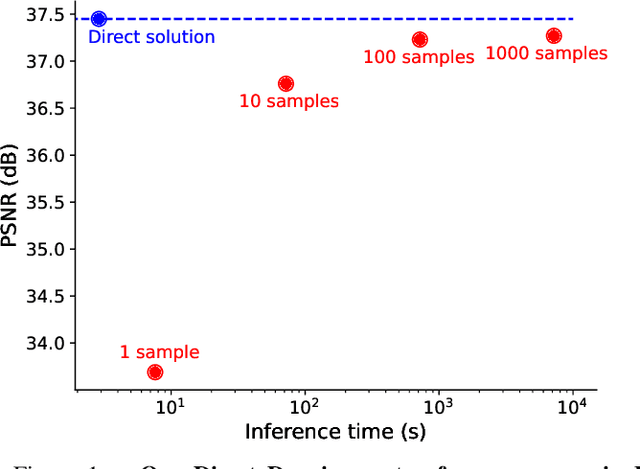
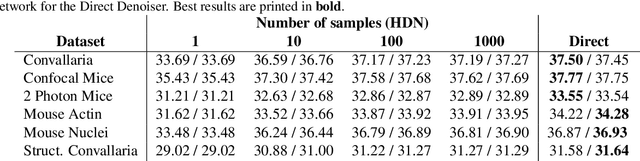
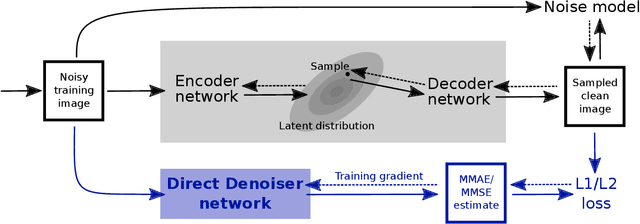
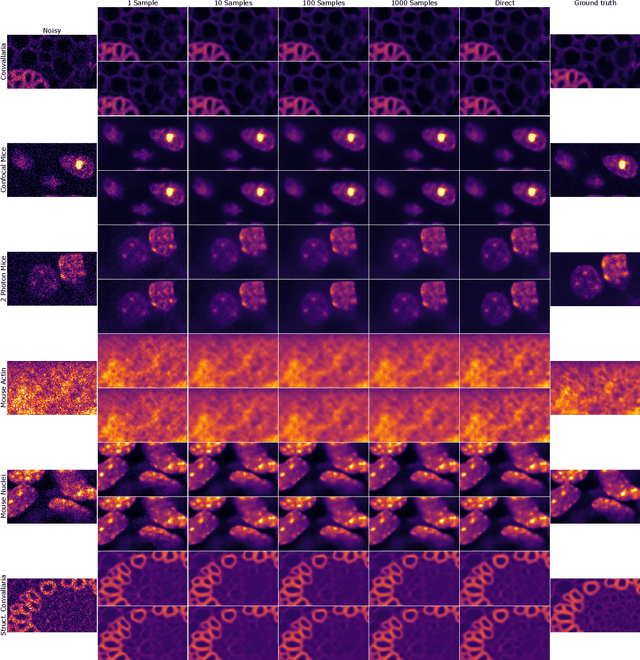
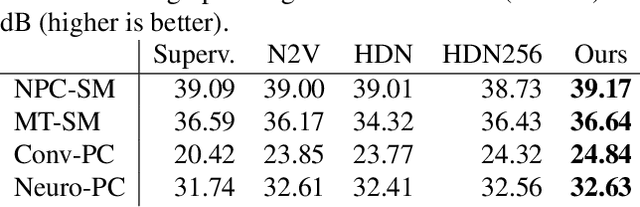
Abstract:Traditional supervised denoisers are trained using pairs of noisy input and clean target images. They learn to predict a central tendency of the posterior distribution over possible clean images. When, e.g., trained with the popular quadratic loss function, the network's output will correspond to the minimum mean square error (MMSE) estimate. Unsupervised denoisers based on Variational AutoEncoders (VAEs) have succeeded in achieving state-of-the-art results while requiring only unpaired noisy data as training input. In contrast to the traditional supervised approach, unsupervised denoisers do not directly produce a single prediction, such as the MMSE estimate, but allow us to draw samples from the posterior distribution of clean solutions corresponding to the noisy input. To approximate the MMSE estimate during inference, unsupervised methods have to create and draw a large number of samples - a computationally expensive process - rendering the approach inapplicable in many situations. Here, we present an alternative approach that trains a deterministic network alongside the VAE to directly predict a central tendency. Our method achieves results that surpass the results achieved by the unsupervised method at a fraction of the computational cost.
Unsupervised Structured Noise Removal with Variational Lossy Autoencoder
Oct 11, 2023Abstract:Most unsupervised denoising methods are based on the assumption that imaging noise is either pixel-independent, i.e., spatially uncorrelated, or signal-independent, i.e., purely additive. However, in practice many imaging setups, especially in microscopy, suffer from a combination of signal-dependent noise (e.g. Poisson shot noise) and axis-aligned correlated noise (e.g. stripe shaped scanning or readout artifacts). In this paper, we present the first unsupervised deep learning-based denoiser that can remove this type of noise without access to any clean images or a noise model. Unlike self-supervised techniques, our method does not rely on removing pixels by masking or subsampling so can utilize all available information. We implement a Variational Autoencoder (VAE) with a specially designed autoregressive decoder capable of modelling the noise component of an image but incapable of independently modelling the underlying clean signal component. As a consequence, our VAE's encoder learns to encode only underlying clean signal content and to discard imaging noise. We also propose an additional decoder for mapping the encoder's latent variables back into image space, thereby sampling denoised images. Experimental results demonstrate that our approach surpasses existing methods for self- and unsupervised image denoising while being robust with respect to the size of the autoregressive receptive field. Code for this project can be found at https://github.com/krulllab/DVLAE.
Image Denoising and the Generative Accumulation of Photons
Aug 01, 2023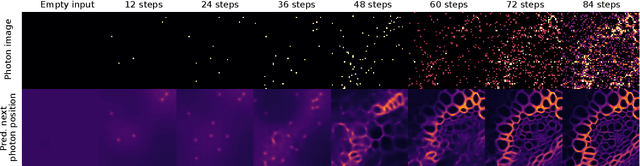
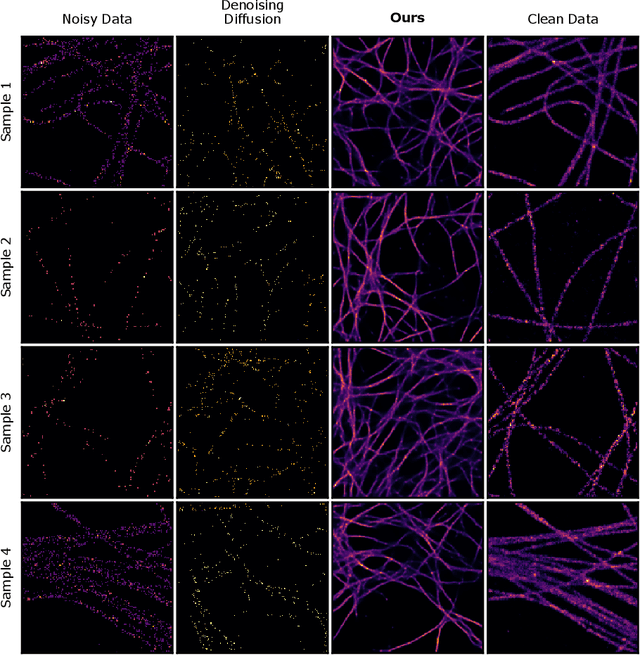
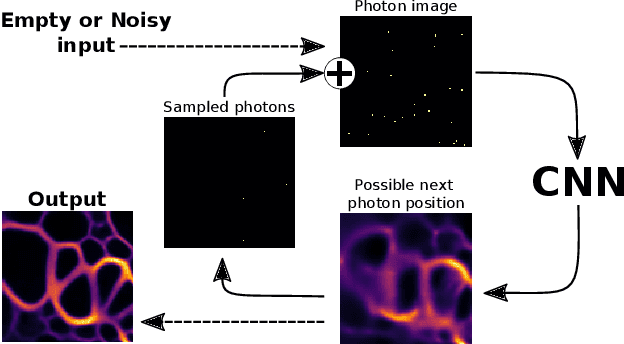
Abstract:We present a fresh perspective on shot noise corrupted images and noise removal. By viewing image formation as the sequential accumulation of photons on a detector grid, we show that a network trained to predict where the next photon could arrive is in fact solving the minimum mean square error (MMSE) denoising task. This new perspective allows us to make three contributions: We present a new strategy for self-supervised denoising, We present a new method for sampling from the posterior of possible solutions by iteratively sampling and adding small numbers of photons to the image. We derive a full generative model by starting this process from an empty canvas. We call this approach generative accumulation of photons (GAP). We evaluate our method quantitatively and qualitatively on 4 new fluorescence microscopy datasets, which will be made available to the community. We find that it outperforms supervised, self-supervised and unsupervised baselines or performs on-par.
μSplit: efficient image decomposition for microscopy data
Nov 23, 2022



Abstract:Light microscopy is routinely used to look at living cells and biological tissues at sub-cellular resolution. Components of the imaged cells can be highlighted using fluorescent labels, allowing biologists to investigate individual structures of interest. Given the complexity of biological processes, it is typically necessary to look at multiple structures simultaneously, typically via a temporal multiplexing scheme. Still, imaging more than 3 or 4 structures in this way is difficult for technical reasons and limits the rate of scientific progress in the life sciences. Hence, a computational method to split apart (decompose) superimposed biological structures acquired in a single image channel, i.e. without temporal multiplexing, would have tremendous impact. Here we present {\mu}Split, a dedicated approach for trained image decomposition. We find that best results using regular deep architectures is achieved when large image patches are used during training, making memory consumption the limiting factor to further improving performance. We therefore introduce lateral contextualization (LC), a memory efficient way to train deep networks that operate well on small input patches. In later layers, additional image context is fed at adequately lowered resolution. We integrate LC with Hierarchical Autoencoders and Hierarchical VAEs.For the latter, we also present a modified ELBO loss and show that it enables sound VAE training. We apply {\mu}Split to five decomposition tasks, one on a synthetic dataset, four others derived from two real microscopy datasets. LC consistently achieves SOTA results, while simultaneously requiring considerably less GPU memory than competing architectures not using LC. When introducing LC, results obtained with the above-mentioned vanilla architectures do on average improve by 2.36 dB (PSNR decibel), with individual improvements ranging from 0.9 to 3.4 dB.
Improving Blind Spot Denoising for Microscopy
Aug 19, 2020



Abstract:Many microscopy applications are limited by the total amount of usable light and are consequently challenged by the resulting levels of noise in the acquired images. This problem is often addressed via (supervised) deep learning based denoising. Recently, by making assumptions about the noise statistics, self-supervised methods have emerged. Such methods are trained directly on the images that are to be denoised and do not require additional paired training data. While achieving remarkable results, self-supervised methods can produce high-frequency artifacts and achieve inferior results compared to supervised approaches. Here we present a novel way to improve the quality of self-supervised denoising. Considering that light microscopy images are usually diffraction-limited, we propose to include this knowledge in the denoising process. We assume the clean image to be the result of a convolution with a point spread function (PSF) and explicitly include this operation at the end of our neural network. As a consequence, we are able to eliminate high-frequency artifacts and achieve self-supervised results that are very close to the ones achieved with traditional supervised methods.
DivNoising: Diversity Denoising with Fully Convolutional Variational Autoencoders
Jun 10, 2020



Abstract:Deep Learning based methods have emerged as the indisputable leaders for virtually all image restoration tasks. Especially in the domain of microscopy images, various content-aware image restoration (CARE) approaches are now used to improve the interpretability of acquired data. But there are limitations to what can be restored in corrupted images, and any given method needs to make a sensible compromise between many possible clean signals when predicting a restored image. Here, we propose DivNoising -- a denoising approach based on fully-convolutional variational autoencoders, overcoming this problem by predicting a whole distribution of denoised images. Our method is unsupervised, requiring only noisy images and a description of the imaging noise, which can be measured or bootstrapped from noisy data. If desired, consensus predictions can be inferred from a set of DivNoising predictions, leading to competitive results with other unsupervised methods and, on occasion, even with the supervised state-of-the-art. DivNoising samples from the posterior enable a plethora of useful applications. We are (i) discussing how optical character recognition (OCR) applications could benefit from diverse predictions on ambiguous data, and (ii) show in detail how instance cell segmentation gains performance when using diverse DivNoising predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge