Samuel Tonks
Can virtual staining for high-throughput screening generalize?
Jul 09, 2024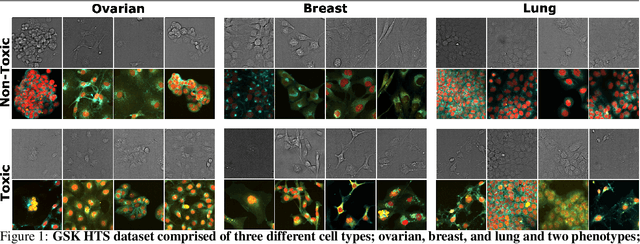
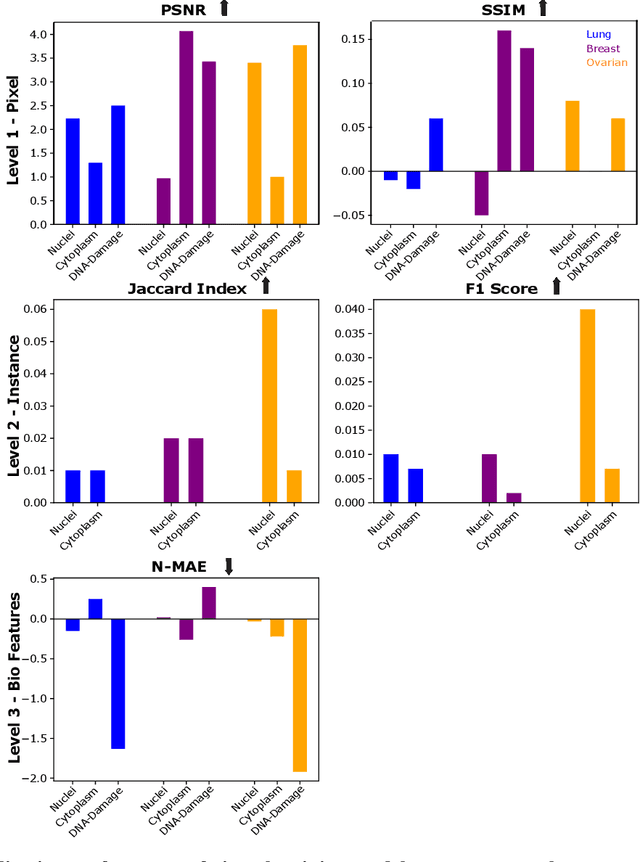
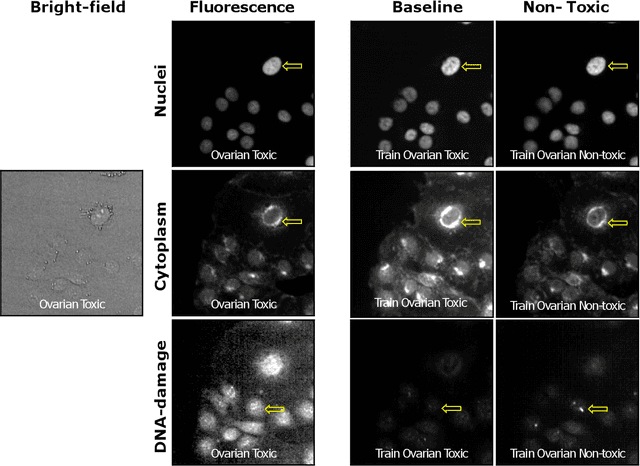
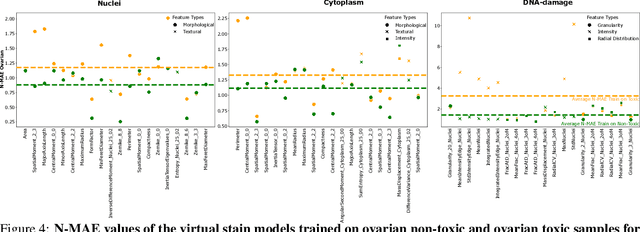
Abstract:The large volume and variety of imaging data from high-throughput screening (HTS) in the pharmaceutical industry present an excellent resource for training virtual staining models. However, the potential of models trained under one set of experimental conditions to generalize to other conditions remains underexplored. This study systematically investigates whether data from three cell types (lung, ovarian, and breast) and two phenotypes (toxic and non-toxic conditions) commonly found in HTS can effectively train virtual staining models to generalize across three typical HTS distribution shifts: unseen phenotypes, unseen cell types, and the combination of both. Utilizing a dataset of 772,416 paired bright-field, cytoplasm, nuclei, and DNA-damage stain images, we evaluate the generalization capabilities of models across pixel-based, instance-wise, and biological-feature-based levels. Our findings indicate that training virtual nuclei and cytoplasm models on non-toxic condition samples not only generalizes to toxic condition samples but leads to improved performance across all evaluation levels compared to training on toxic condition samples. Generalization to unseen cell types shows variability depending on the cell type; models trained on ovarian or lung cell samples often perform well under other conditions, while those trained on breast cell samples consistently show poor generalization. Generalization to unseen cell types and phenotypes shows good generalization across all levels of evaluation compared to addressing unseen cell types alone. This study represents the first large-scale, data-centric analysis of the generalization capability of virtual staining models trained on diverse HTS datasets, providing valuable strategies for experimental training data generation.
Image Denoising and the Generative Accumulation of Photons
Aug 01, 2023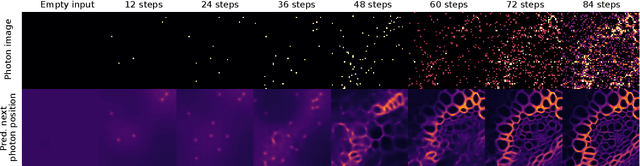
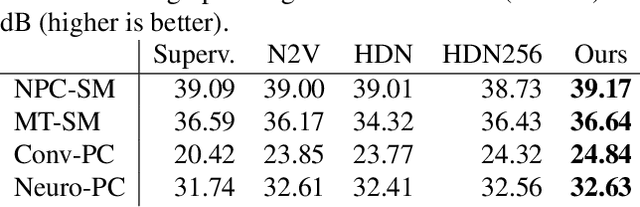
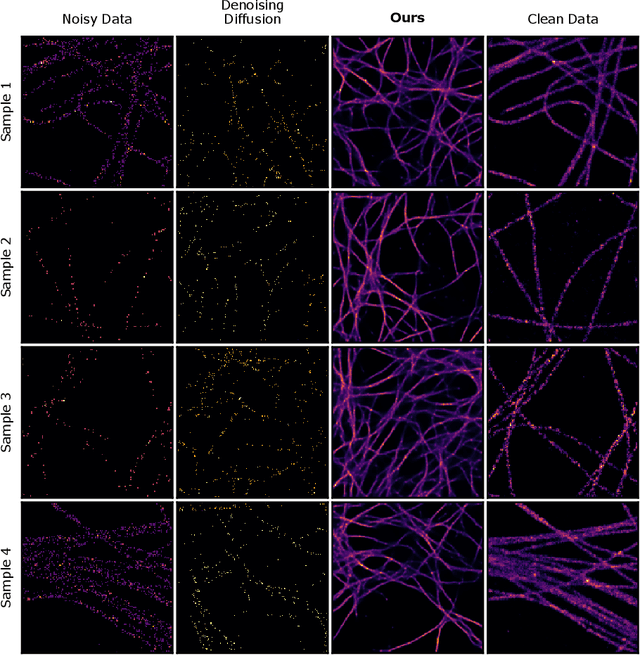
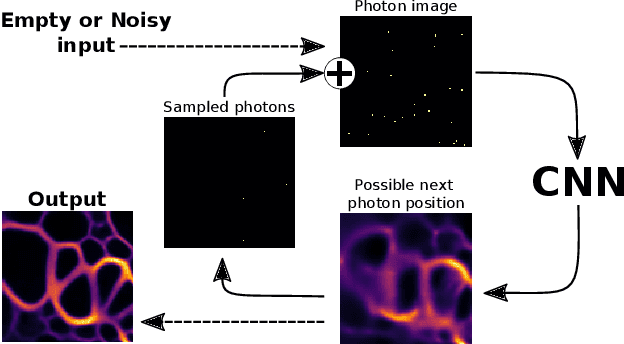
Abstract:We present a fresh perspective on shot noise corrupted images and noise removal. By viewing image formation as the sequential accumulation of photons on a detector grid, we show that a network trained to predict where the next photon could arrive is in fact solving the minimum mean square error (MMSE) denoising task. This new perspective allows us to make three contributions: We present a new strategy for self-supervised denoising, We present a new method for sampling from the posterior of possible solutions by iteratively sampling and adding small numbers of photons to the image. We derive a full generative model by starting this process from an empty canvas. We call this approach generative accumulation of photons (GAP). We evaluate our method quantitatively and qualitatively on 4 new fluorescence microscopy datasets, which will be made available to the community. We find that it outperforms supervised, self-supervised and unsupervised baselines or performs on-par.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge