Tim-Oliver Buchholz
N2V2 -- Fixing Noise2Void Checkerboard Artifacts with Modified Sampling Strategies and a Tweaked Network Architecture
Nov 21, 2022



Abstract:In recent years, neural network based image denoising approaches have revolutionized the analysis of biomedical microscopy data. Self-supervised methods, such as Noise2Void (N2V), are applicable to virtually all noisy datasets, even without dedicated training data being available. Arguably, this facilitated the fast and widespread adoption of N2V throughout the life sciences. Unfortunately, the blind-spot training underlying N2V can lead to rather visible checkerboard artifacts, thereby reducing the quality of final predictions considerably. In this work, we present two modifications to the vanilla N2V setup that both help to reduce the unwanted artifacts considerably. Firstly, we propose a modified network architecture, i.e., using BlurPool instead of MaxPool layers throughout the used U-Net, rolling back the residual U-Net to a non-residual U-Net, and eliminating the skip connections at the uppermost U-Net level. Additionally, we propose new replacement strategies to determine the pixel intensity values that fill in the elected blind-spot pixels. We validate our modifications on a range of microscopy and natural image data. Based on added synthetic noise from multiple noise types and at varying amplitudes, we show that both proposed modifications push the current state-of-the-art for fully self-supervised image denoising.
Fourier Image Transformer
May 03, 2021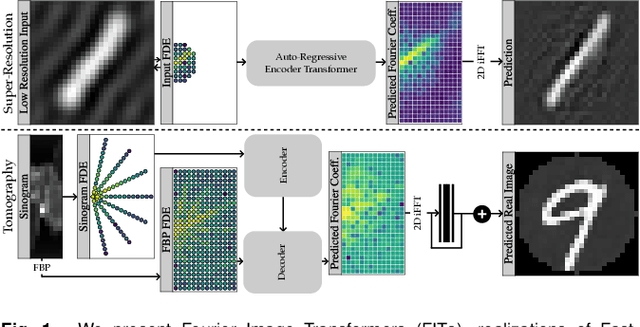
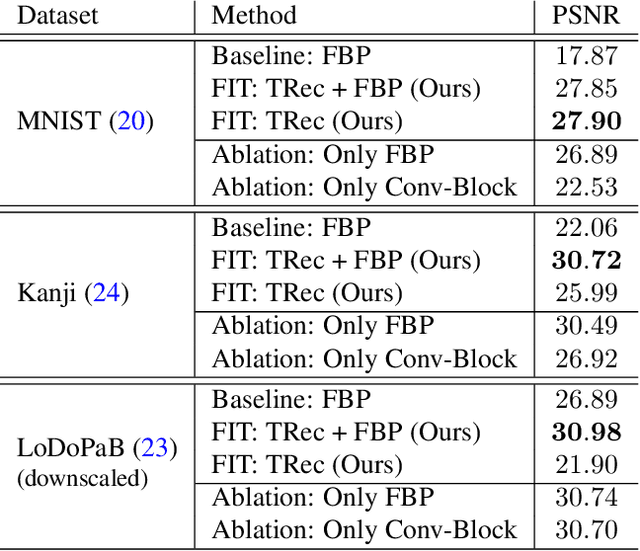
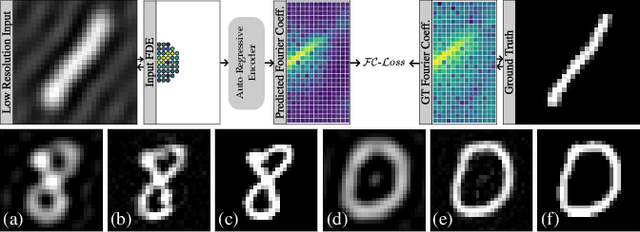
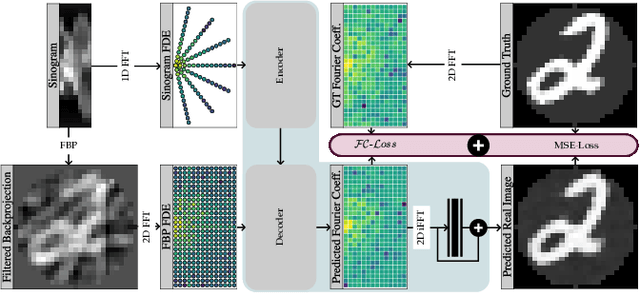
Abstract:Transformer architectures show spectacular performance on NLP tasks and have recently also been used for tasks such as image completion or image classification. Here we propose to use a sequential image representation, where each prefix of the complete sequence describes the whole image at reduced resolution. Using such Fourier Domain Encodings (FDEs), an auto-regressive image completion task is equivalent to predicting a higher resolution output given a low-resolution input. Additionally, we show that an encoder-decoder setup can be used to query arbitrary Fourier coefficients given a set of Fourier domain observations. We demonstrate the practicality of this approach in the context of computed tomography (CT) image reconstruction. In summary, we show that Fourier Image Transformer (FIT) can be used to solve relevant image analysis tasks in Fourier space, a domain inherently inaccessible to convolutional architectures.
DenoiSeg: Joint Denoising and Segmentation
May 06, 2020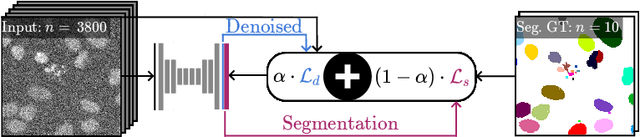
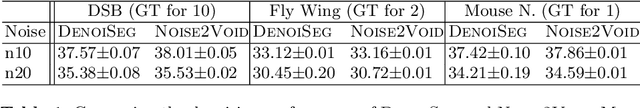
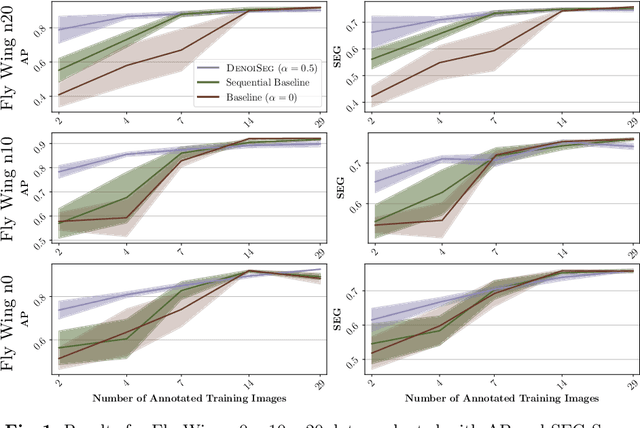
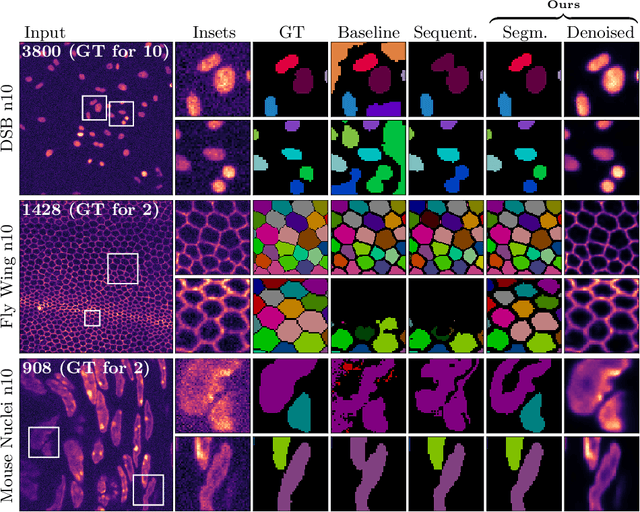
Abstract:Microscopy image analysis often requires the segmentation of objects, but training data for this task is typically scarce and hard to obtain. Here we propose DenoiSeg, a new method that can be trained end-to-end on only a few annotated ground truth segmentations. We achieve this by extending Noise2Void, a self-supervised denoising scheme that can be trained on noisy images alone, to also predict dense 3-class segmentations. The reason for the success of our method is that segmentation can profit from denoising, especially when performed jointly within the same network. The network becomes a denoising expert by seeing all available raw data, while co-learning to segment, even if only a few segmentation labels are available. This hypothesis is additionally fueled by our observation that the best segmentation results on high quality (very low noise) raw data are obtained when moderate amounts of synthetic noise are added. This renders the denoising-task non-trivial and unleashes the desired co-learning effect. We believe that DenoiSeg offers a viable way to circumvent the tremendous hunger for high quality training data and effectively enables few-shot learning of dense segmentations.
Leveraging Self-supervised Denoising for Image Segmentation
Dec 13, 2019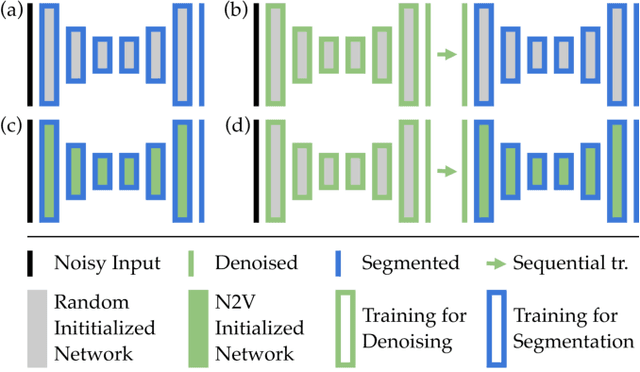
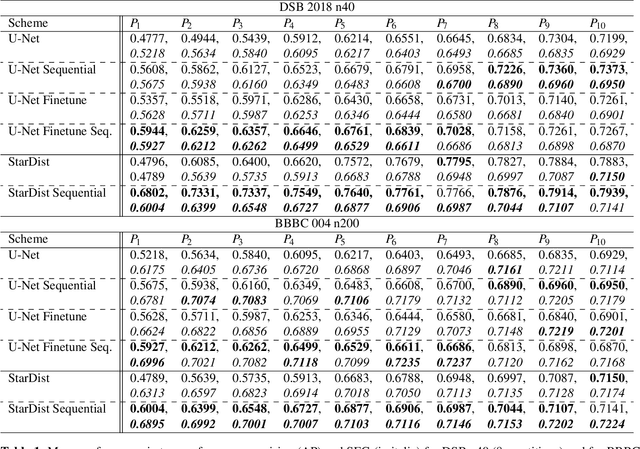
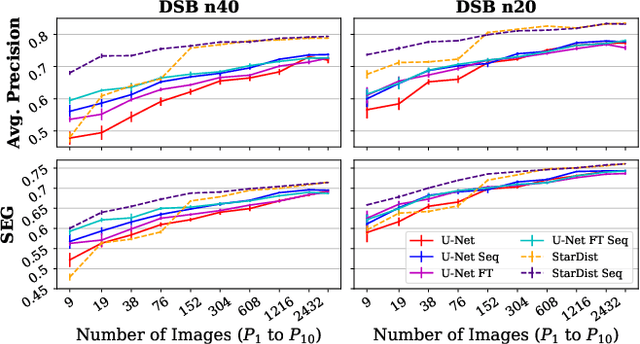
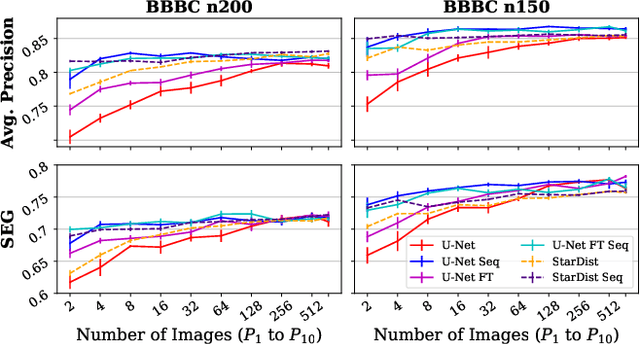
Abstract:Deep learning (DL) has arguably emerged as the method of choice for the detection and segmentation of biological structures in microscopy images. However, DL typically needs copious amounts of annotated training data that is for biomedical projects typically not available and excessively expensive to generate. Additionally, tasks become harder in the presence of noise, requiring even more high-quality training data. Hence, we propose to use denoising networks to improve the performance of other DL-based image segmentation methods. More specifically, we present ideas on how state-of-the-art self-supervised CARE networks can improve cell/nuclei segmentation in microscopy data. Using two state-of-the-art baseline methods, U-Net and StarDist, we show that our ideas consistently improve the quality of resulting segmentations, especially when only limited training data for noisy micrographs are available.
Noise2Void - Learning Denoising from Single Noisy Images
Nov 27, 2018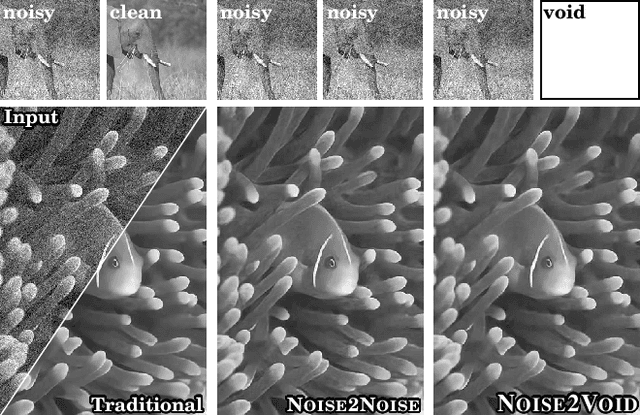
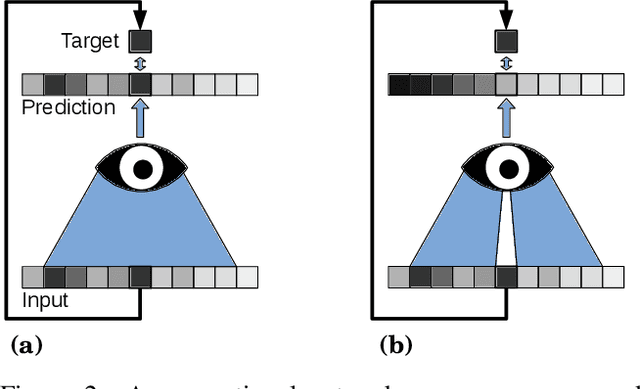
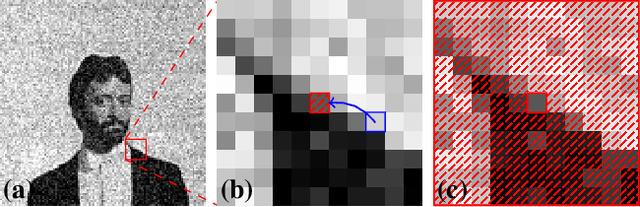
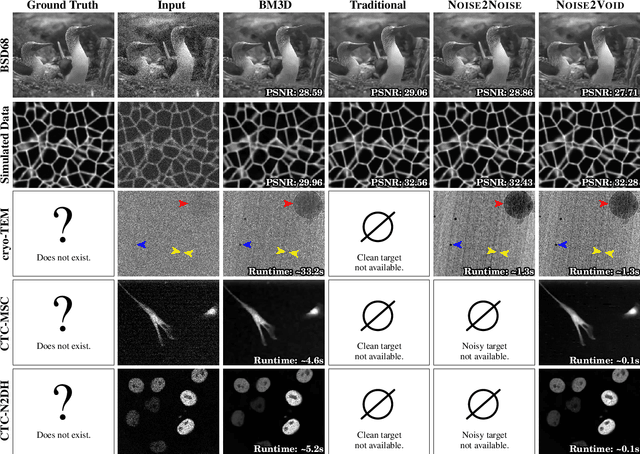
Abstract:The field of image denoising is currently dominated by discriminative deep learning methods that are trained on pairs of noisy input and clean target images. Recently it has been shown that such methods can also be trained without clean targets. Instead, independent pairs of noisy images can be used, in an approach known as Noise2Noise (N2N). Here, we introduce Noise2Void (N2V), a training scheme that takes this idea one step further. It does not require noisy image pairs, nor clean target images. Consequently, N2V allows us to train directly on the body of data to be denoised and can therefore be applied when other methods cannot. Especially interesting is the application to biomedical image data, where the acquisition of training targets, clean or noisy, is frequently not possible. We compare the performance of N2V to approaches that have either clean target images and/or noisy image pairs available. Intuitively, N2V cannot be expected to outperform methods that have more information available during training. Still, we observe that the denoising performance of Noise2Void drops in moderation and compares favorably to training-free denoising methods.
Cryo-CARE: Content-Aware Image Restoration for Cryo-Transmission Electron Microscopy Data
Oct 15, 2018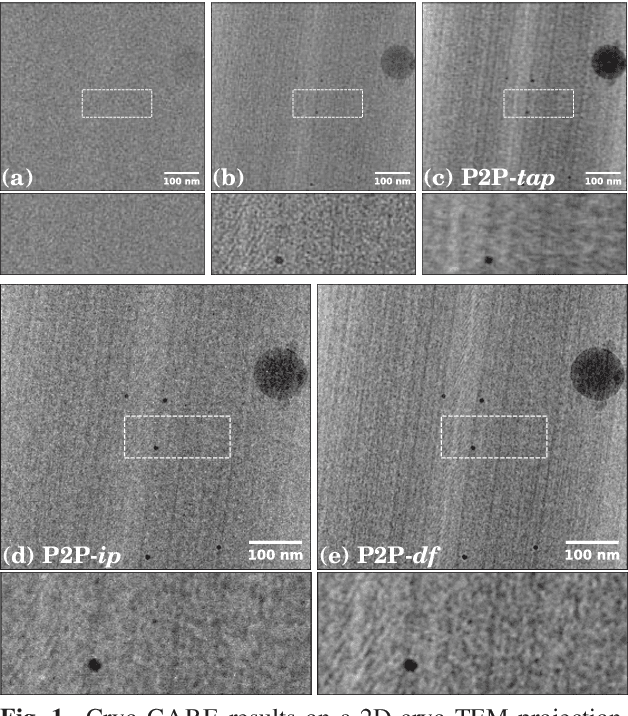
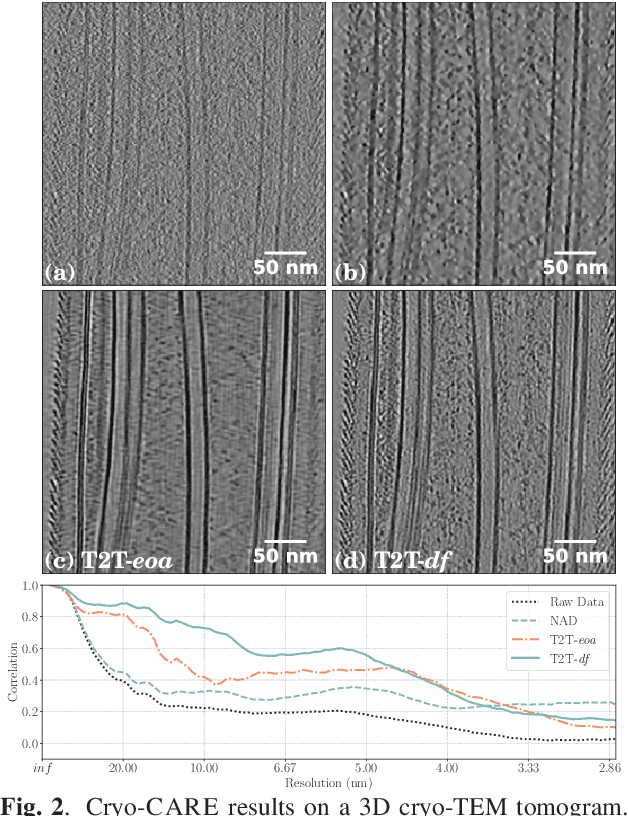
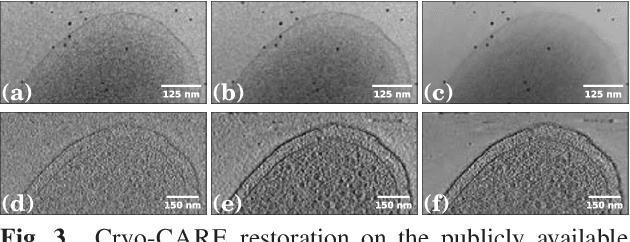
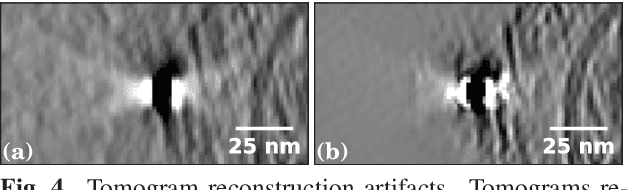
Abstract:Multiple approaches to use deep learning for image restoration have recently been proposed. Training such approaches requires well registered pairs of high and low quality images. While this is easily achievable for many imaging modalities, e.g. fluorescence light microscopy, for others it is not. Cryo-transmission electron microscopy (cryo-TEM) could profoundly benefit from improved denoising methods, unfortunately it is one of the latter. Here we show how recent advances in network training for image restoration tasks, i.e. denoising, can be applied to cryo-TEM data. We describe our proposed method and show how it can be applied to single cryo-TEM projections and whole cryo-tomographic image volumes. Our proposed restoration method dramatically increases contrast in cryo-TEM images, which improves the interpretability of the acquired data. Furthermore we show that automated downstream processing on restored image data, demonstrated on a dense segmentation task, leads to improved results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge