Alexander Freytag
Carl Zeiss AG
Prompt-Tuning SAM: From Generalist to Specialist with only 2048 Parameters and 16 Training Images
Apr 23, 2025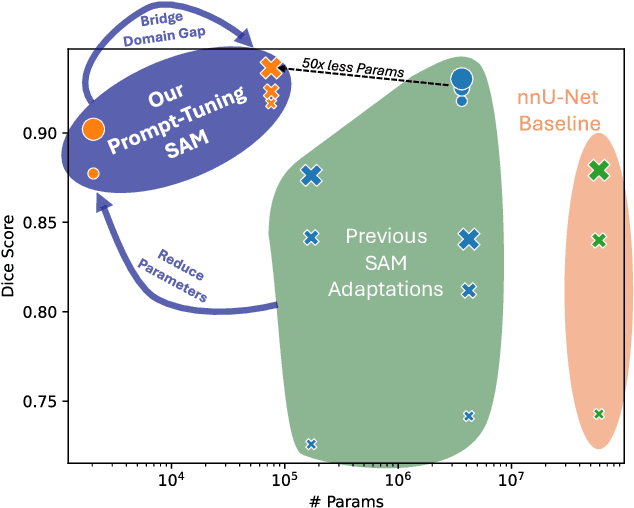
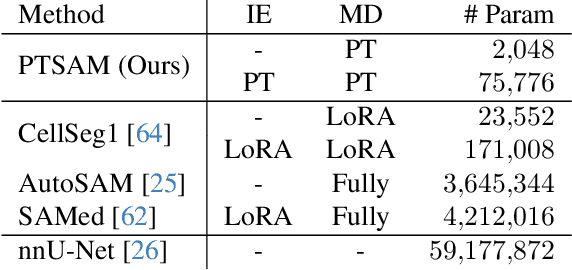
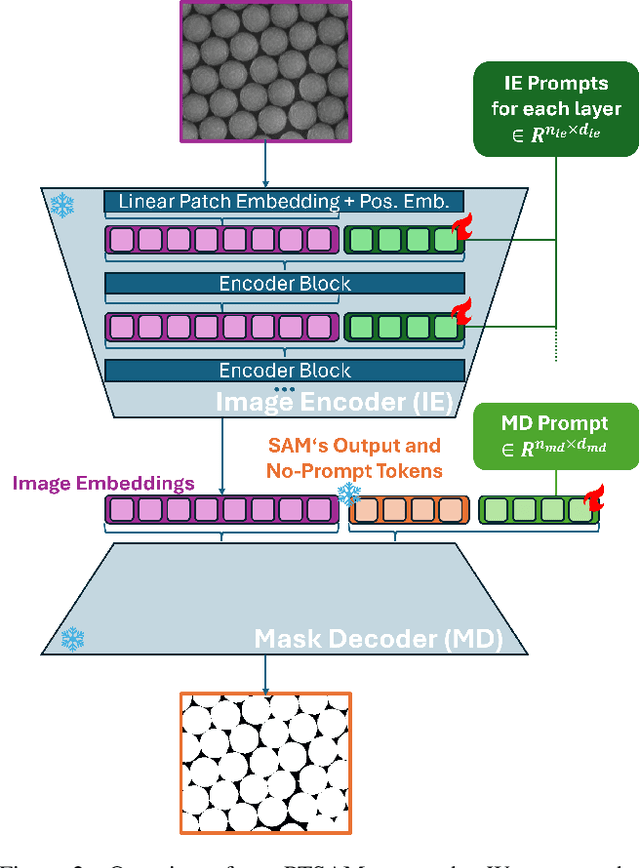
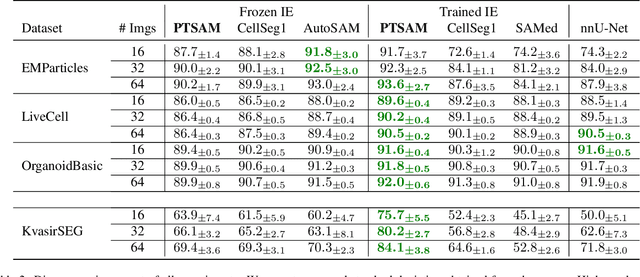
Abstract:The Segment Anything Model (SAM) is widely used for segmenting a diverse range of objects in natural images from simple user prompts like points or bounding boxes. However, SAM's performance decreases substantially when applied to non-natural domains like microscopic imaging. Furthermore, due to SAM's interactive design, it requires a precise prompt for each image and object, which is unfeasible in many automated biomedical applications. Previous solutions adapt SAM by training millions of parameters via fine-tuning large parts of the model or of adapter layers. In contrast, we show that as little as 2,048 additional parameters are sufficient for turning SAM into a use-case specialist for a certain downstream task. Our novel PTSAM (prompt-tuned SAM) method uses prompt-tuning, a parameter-efficient fine-tuning technique, to adapt SAM for a specific task. We validate the performance of our approach on multiple microscopic and one medical dataset. Our results show that prompt-tuning only SAM's mask decoder already leads to a performance on-par with state-of-the-art techniques while requiring roughly 2,000x less trainable parameters. For addressing domain gaps, we find that additionally prompt-tuning SAM's image encoder is beneficial, further improving segmentation accuracy by up to 18% over state-of-the-art results. Since PTSAM can be reliably trained with as little as 16 annotated images, we find it particularly helpful for applications with limited training data and domain shifts.
Intra-operative Brain Tumor Detection with Deep Learning-Optimized Hyperspectral Imaging
Feb 06, 2023Abstract:Surgery for gliomas (intrinsic brain tumors), especially when low-grade, is challenging due to the infiltrative nature of the lesion. Currently, no real-time, intra-operative, label-free and wide-field tool is available to assist and guide the surgeon to find the relevant demarcations for these tumors. While marker-based methods exist for the high-grade glioma case, there is no convenient solution available for the low-grade case; thus, marker-free optical techniques represent an attractive option. Although RGB imaging is a standard tool in surgical microscopes, it does not contain sufficient information for tissue differentiation. We leverage the richer information from hyperspectral imaging (HSI), acquired with a snapscan camera in the 468-787 nm range, coupled to a surgical microscope, to build a deep-learning-based diagnostic tool for cancer resection with potential for intra-operative guidance. However, the main limitation of the HSI snapscan camera is the image acquisition time, limiting its widespread deployment in the operation theater. Here, we investigate the effect of HSI channel reduction and pre-selection to scope the design space for the development of cheaper and faster sensors. Neural networks are used to identify the most important spectral channels for tumor tissue differentiation, optimizing the trade-off between the number of channels and precision to enable real-time intra-surgical application. We evaluate the performance of our method on a clinical dataset that was acquired during surgery on five patients. By demonstrating the possibility to efficiently detect low-grade glioma, these results can lead to better cancer resection demarcations, potentially improving treatment effectiveness and patient outcome.
N2V2 -- Fixing Noise2Void Checkerboard Artifacts with Modified Sampling Strategies and a Tweaked Network Architecture
Nov 21, 2022



Abstract:In recent years, neural network based image denoising approaches have revolutionized the analysis of biomedical microscopy data. Self-supervised methods, such as Noise2Void (N2V), are applicable to virtually all noisy datasets, even without dedicated training data being available. Arguably, this facilitated the fast and widespread adoption of N2V throughout the life sciences. Unfortunately, the blind-spot training underlying N2V can lead to rather visible checkerboard artifacts, thereby reducing the quality of final predictions considerably. In this work, we present two modifications to the vanilla N2V setup that both help to reduce the unwanted artifacts considerably. Firstly, we propose a modified network architecture, i.e., using BlurPool instead of MaxPool layers throughout the used U-Net, rolling back the residual U-Net to a non-residual U-Net, and eliminating the skip connections at the uppermost U-Net level. Additionally, we propose new replacement strategies to determine the pixel intensity values that fill in the elected blind-spot pixels. We validate our modifications on a range of microscopy and natural image data. Based on added synthetic noise from multiple noise types and at varying amplitudes, we show that both proposed modifications push the current state-of-the-art for fully self-supervised image denoising.
Every Annotation Counts: Multi-label Deep Supervision for Medical Image Segmentation
Apr 27, 2021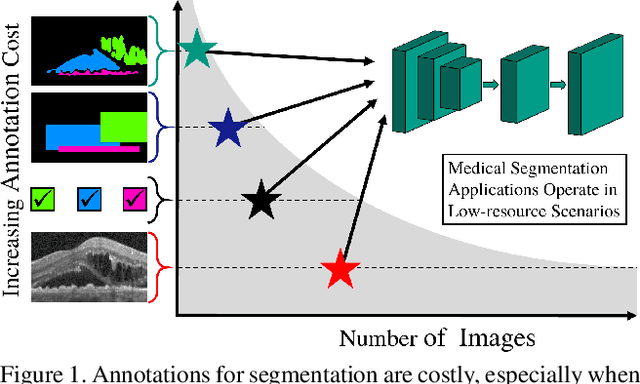
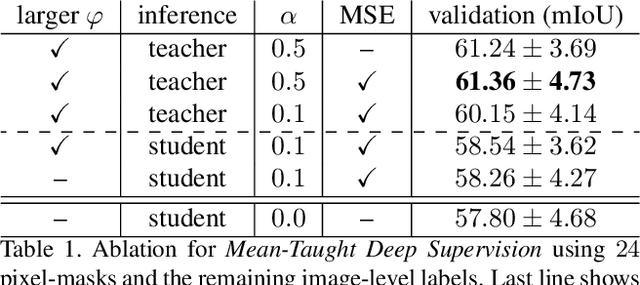
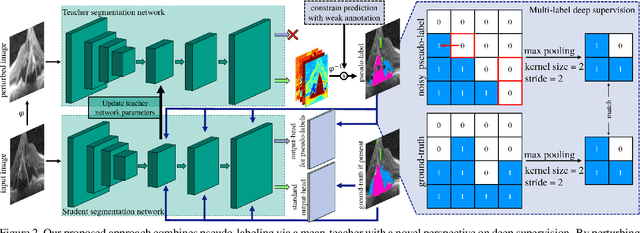
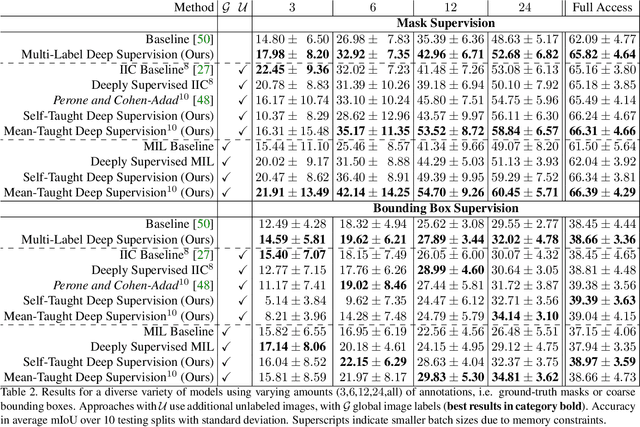
Abstract:Pixel-wise segmentation is one of the most data and annotation hungry tasks in our field. Providing representative and accurate annotations is often mission-critical especially for challenging medical applications. In this paper, we propose a semi-weakly supervised segmentation algorithm to overcome this barrier. Our approach is based on a new formulation of deep supervision and student-teacher model and allows for easy integration of different supervision signals. In contrast to previous work, we show that care has to be taken how deep supervision is integrated in lower layers and we present multi-label deep supervision as the most important secret ingredient for success. With our novel training regime for segmentation that flexibly makes use of images that are either fully labeled, marked with bounding boxes, just global labels, or not at all, we are able to cut the requirement for expensive labels by 94.22% - narrowing the gap to the best fully supervised baseline to only 5% mean IoU. Our approach is validated by extensive experiments on retinal fluid segmentation and we provide an in-depth analysis of the anticipated effect each annotation type can have in boosting segmentation performance.
Active and Continuous Exploration with Deep Neural Networks and Expected Model Output Changes
Dec 19, 2016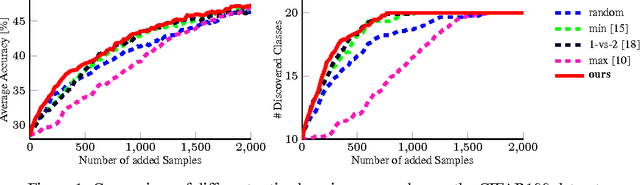
Abstract:The demands on visual recognition systems do not end with the complexity offered by current large-scale image datasets, such as ImageNet. In consequence, we need curious and continuously learning algorithms that actively acquire knowledge about semantic concepts which are present in available unlabeled data. As a step towards this goal, we show how to perform continuous active learning and exploration, where an algorithm actively selects relevant batches of unlabeled examples for annotation. These examples could either belong to already known or to yet undiscovered classes. Our algorithm is based on a new generalization of the Expected Model Output Change principle for deep architectures and is especially tailored to deep neural networks. Furthermore, we show easy-to-implement approximations that yield efficient techniques for active selection. Empirical experiments show that our method outperforms currently used heuristics.
Seeing through bag-of-visual-word glasses: towards understanding quantization effects in feature extraction methods
Aug 20, 2014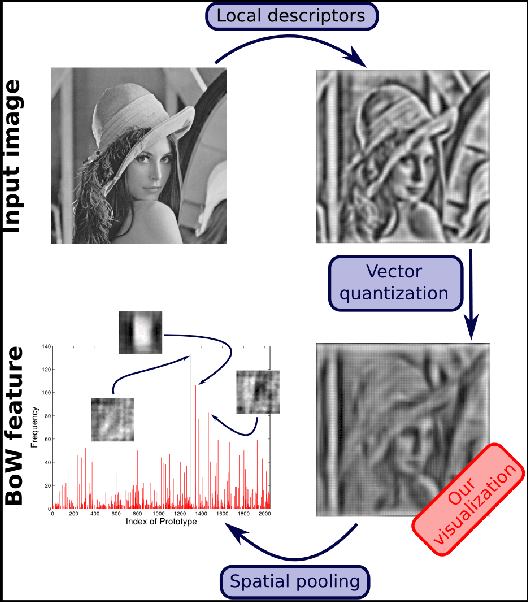
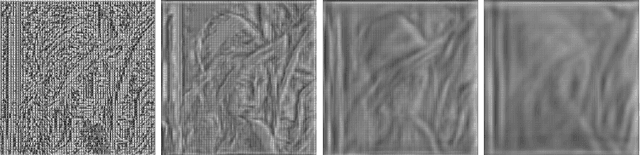
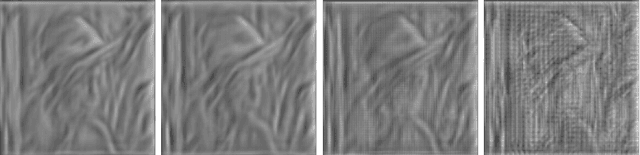

Abstract:Vector-quantized local features frequently used in bag-of-visual-words approaches are the backbone of popular visual recognition systems due to both their simplicity and their performance. Despite their success, bag-of-words-histograms basically contain low-level image statistics (e.g., number of edges of different orientations). The question remains how much visual information is "lost in quantization" when mapping visual features to code words? To answer this question, we present an in-depth analysis of the effect of local feature quantization on human recognition performance. Our analysis is based on recovering the visual information by inverting quantized local features and presenting these visualizations with different codebook sizes to human observers. Although feature inversion techniques are around for quite a while, to the best of our knowledge, our technique is the first visualizing especially the effect of feature quantization. Thereby, we are now able to compare single steps in common image classification pipelines to human counterparts.
Fine-grained Categorization -- Short Summary of our Entry for the ImageNet Challenge 2012
Oct 17, 2013
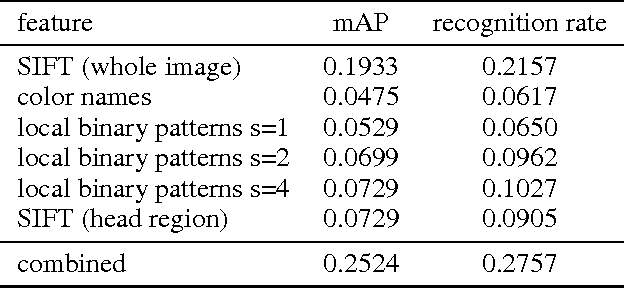
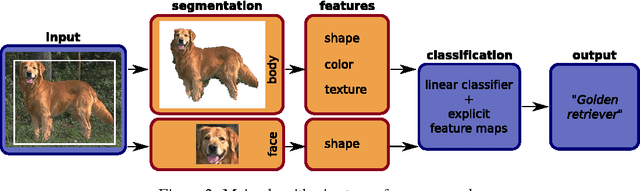
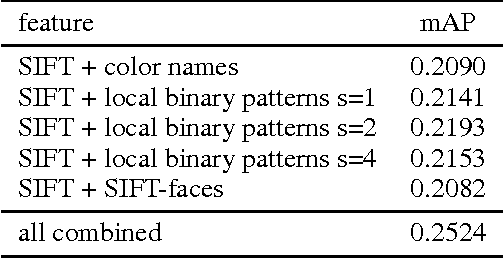
Abstract:In this paper, we tackle the problem of visual categorization of dog breeds, which is a surprisingly challenging task due to simultaneously present low interclass distances and high intra-class variances. Our approach combines several techniques well known in our community but often not utilized for fine-grained recognition: (1) automatic segmentation, (2) efficient part detection, and (3) combination of multiple features. In particular, we demonstrate that a simple head detector embedded in an off-the-shelf recognition pipeline can improve recognition accuracy quite significantly, highlighting the importance of part features for fine-grained recognition tasks. Using our approach, we achieved a 24.59% mean average precision performance on the Stanford dog dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge