Anna Alperovich
Carl Zeiss AG
Robust Tumor Segmentation with Hyperspectral Imaging and Graph Neural Networks
Nov 20, 2023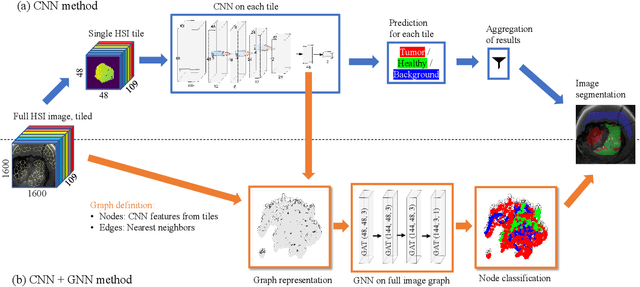
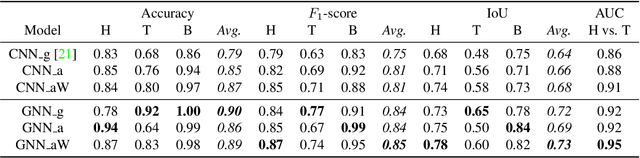
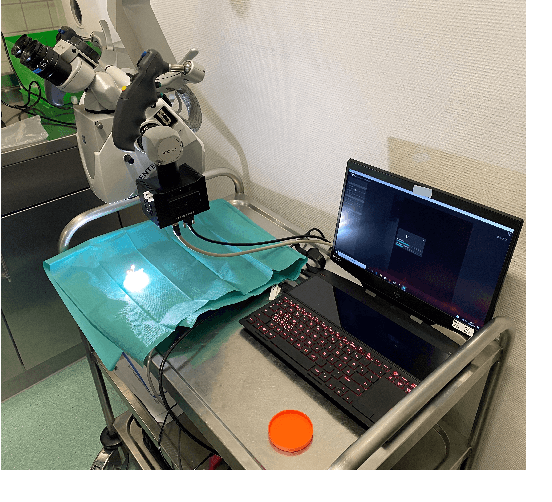
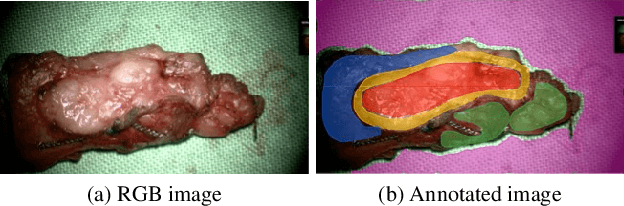
Abstract:Segmenting the boundary between tumor and healthy tissue during surgical cancer resection poses a significant challenge. In recent years, Hyperspectral Imaging (HSI) combined with Machine Learning (ML) has emerged as a promising solution. However, due to the extensive information contained within the spectral domain, most ML approaches primarily classify individual HSI (super-)pixels, or tiles, without taking into account their spatial context. In this paper, we propose an improved methodology that leverages the spatial context of tiles for more robust and smoother segmentation. To address the irregular shapes of tiles, we utilize Graph Neural Networks (GNNs) to propagate context information across neighboring regions. The features for each tile within the graph are extracted using a Convolutional Neural Network (CNN), which is trained simultaneously with the subsequent GNN. Moreover, we incorporate local image quality metrics into the loss function to enhance the training procedure's robustness against low-quality regions in the training images. We demonstrate the superiority of our proposed method using a clinical ex vivo dataset consisting of 51 HSI images from 30 patients. Despite the limited dataset, the GNN-based model significantly outperforms context-agnostic approaches, accurately distinguishing between healthy and tumor tissues, even in images from previously unseen patients. Furthermore, we show that our carefully designed loss function, accounting for local image quality, results in additional improvements. Our findings demonstrate that context-aware GNN algorithms can robustly find tumor demarcations on HSI images, ultimately contributing to better surgery success and patient outcome.
Intra-operative Brain Tumor Detection with Deep Learning-Optimized Hyperspectral Imaging
Feb 06, 2023Abstract:Surgery for gliomas (intrinsic brain tumors), especially when low-grade, is challenging due to the infiltrative nature of the lesion. Currently, no real-time, intra-operative, label-free and wide-field tool is available to assist and guide the surgeon to find the relevant demarcations for these tumors. While marker-based methods exist for the high-grade glioma case, there is no convenient solution available for the low-grade case; thus, marker-free optical techniques represent an attractive option. Although RGB imaging is a standard tool in surgical microscopes, it does not contain sufficient information for tissue differentiation. We leverage the richer information from hyperspectral imaging (HSI), acquired with a snapscan camera in the 468-787 nm range, coupled to a surgical microscope, to build a deep-learning-based diagnostic tool for cancer resection with potential for intra-operative guidance. However, the main limitation of the HSI snapscan camera is the image acquisition time, limiting its widespread deployment in the operation theater. Here, we investigate the effect of HSI channel reduction and pre-selection to scope the design space for the development of cheaper and faster sensors. Neural networks are used to identify the most important spectral channels for tumor tissue differentiation, optimizing the trade-off between the number of channels and precision to enable real-time intra-surgical application. We evaluate the performance of our method on a clinical dataset that was acquired during surgery on five patients. By demonstrating the possibility to efficiently detect low-grade glioma, these results can lead to better cancer resection demarcations, potentially improving treatment effectiveness and patient outcome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge