Felix Holm
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
ProtoFlow: Interpretable and Robust Surgical Workflow Modeling with Learned Dynamic Scene Graph Prototypes
Dec 16, 2025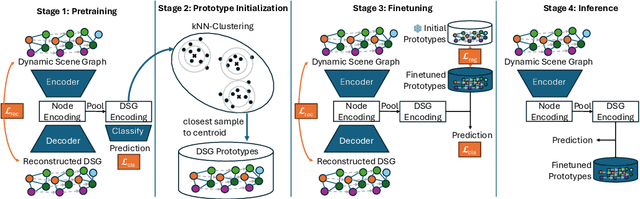
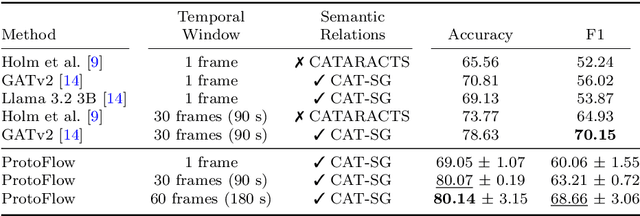
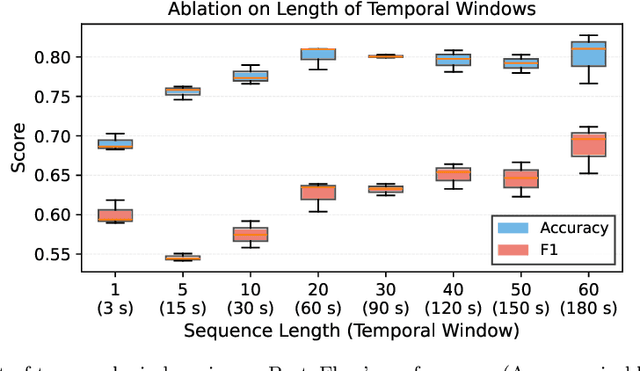
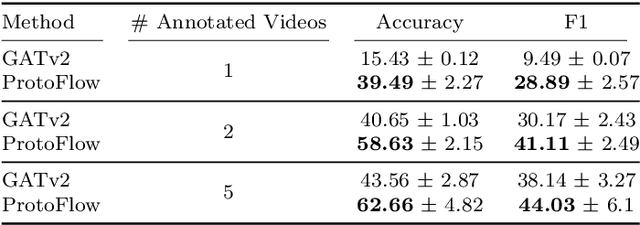
Abstract:Purpose: Detailed surgical recognition is critical for advancing AI-assisted surgery, yet progress is hampered by high annotation costs, data scarcity, and a lack of interpretable models. While scene graphs offer a structured abstraction of surgical events, their full potential remains untapped. In this work, we introduce ProtoFlow, a novel framework that learns dynamic scene graph prototypes to model complex surgical workflows in an interpretable and robust manner. Methods: ProtoFlow leverages a graph neural network (GNN) encoder-decoder architecture that combines self-supervised pretraining for rich representation learning with a prototype-based fine-tuning stage. This process discovers and refines core prototypes that encapsulate recurring, clinically meaningful patterns of surgical interaction, forming an explainable foundation for workflow analysis. Results: We evaluate our approach on the fine-grained CAT-SG dataset. ProtoFlow not only outperforms standard GNN baselines in overall accuracy but also demonstrates exceptional robustness in limited-data, few-shot scenarios, maintaining strong performance when trained on as few as one surgical video. Our qualitative analyses further show that the learned prototypes successfully identify distinct surgical sub-techniques and provide clear, interpretable insights into workflow deviations and rare complications. Conclusion: By uniting robust representation learning with inherent explainability, ProtoFlow represents a significant step toward developing more transparent, reliable, and data-efficient AI systems, accelerating their potential for clinical adoption in surgical training, real-time decision support, and workflow optimization.
SANGRIA: Surgical Video Scene Graph Optimization for Surgical Workflow Prediction
Jul 29, 2024Abstract:Graph-based holistic scene representations facilitate surgical workflow understanding and have recently demonstrated significant success. However, this task is often hindered by the limited availability of densely annotated surgical scene data. In this work, we introduce an end-to-end framework for the generation and optimization of surgical scene graphs on a downstream task. Our approach leverages the flexibility of graph-based spectral clustering and the generalization capability of foundation models to generate unsupervised scene graphs with learnable properties. We reinforce the initial spatial graph with sparse temporal connections using local matches between consecutive frames to predict temporally consistent clusters across a temporal neighborhood. By jointly optimizing the spatiotemporal relations and node features of the dynamic scene graph with the downstream task of phase segmentation, we address the costly and annotation-burdensome task of semantic scene comprehension and scene graph generation in surgical videos using only weak surgical phase labels. Further, by incorporating effective intermediate scene representation disentanglement steps within the pipeline, our solution outperforms the SOTA on the CATARACTS dataset by 8% accuracy and 10% F1 score in surgical workflow recognition
Robust Tumor Segmentation with Hyperspectral Imaging and Graph Neural Networks
Nov 20, 2023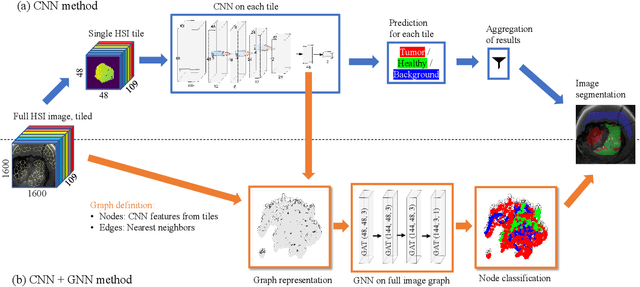
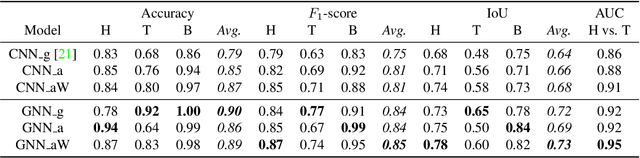
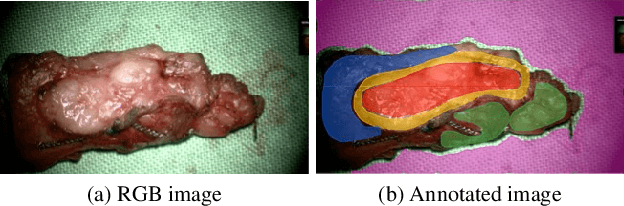
Abstract:Segmenting the boundary between tumor and healthy tissue during surgical cancer resection poses a significant challenge. In recent years, Hyperspectral Imaging (HSI) combined with Machine Learning (ML) has emerged as a promising solution. However, due to the extensive information contained within the spectral domain, most ML approaches primarily classify individual HSI (super-)pixels, or tiles, without taking into account their spatial context. In this paper, we propose an improved methodology that leverages the spatial context of tiles for more robust and smoother segmentation. To address the irregular shapes of tiles, we utilize Graph Neural Networks (GNNs) to propagate context information across neighboring regions. The features for each tile within the graph are extracted using a Convolutional Neural Network (CNN), which is trained simultaneously with the subsequent GNN. Moreover, we incorporate local image quality metrics into the loss function to enhance the training procedure's robustness against low-quality regions in the training images. We demonstrate the superiority of our proposed method using a clinical ex vivo dataset consisting of 51 HSI images from 30 patients. Despite the limited dataset, the GNN-based model significantly outperforms context-agnostic approaches, accurately distinguishing between healthy and tumor tissues, even in images from previously unseen patients. Furthermore, we show that our carefully designed loss function, accounting for local image quality, results in additional improvements. Our findings demonstrate that context-aware GNN algorithms can robustly find tumor demarcations on HSI images, ultimately contributing to better surgery success and patient outcome.
Dynamic Scene Graph Representation for Surgical Video
Sep 25, 2023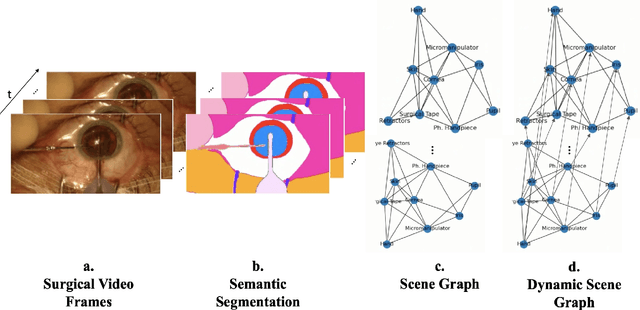
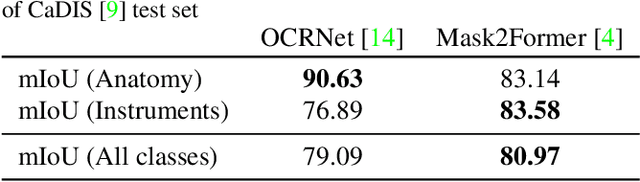
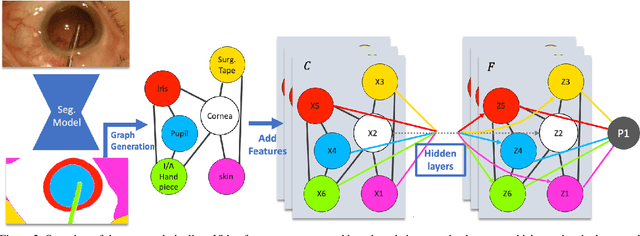
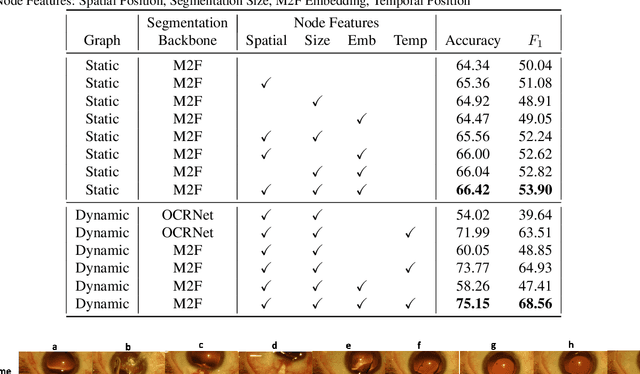
Abstract:Surgical videos captured from microscopic or endoscopic imaging devices are rich but complex sources of information, depicting different tools and anatomical structures utilized during an extended amount of time. Despite containing crucial workflow information and being commonly recorded in many procedures, usage of surgical videos for automated surgical workflow understanding is still limited. In this work, we exploit scene graphs as a more holistic, semantically meaningful and human-readable way to represent surgical videos while encoding all anatomical structures, tools, and their interactions. To properly evaluate the impact of our solutions, we create a scene graph dataset from semantic segmentations from the CaDIS and CATARACTS datasets. We demonstrate that scene graphs can be leveraged through the use of graph convolutional networks (GCNs) to tackle surgical downstream tasks such as surgical workflow recognition with competitive performance. Moreover, we demonstrate the benefits of surgical scene graphs regarding the explainability and robustness of model decisions, which are crucial in the clinical setting.
Whether and When does Endoscopy Domain Pretraining Make Sense?
Mar 30, 2023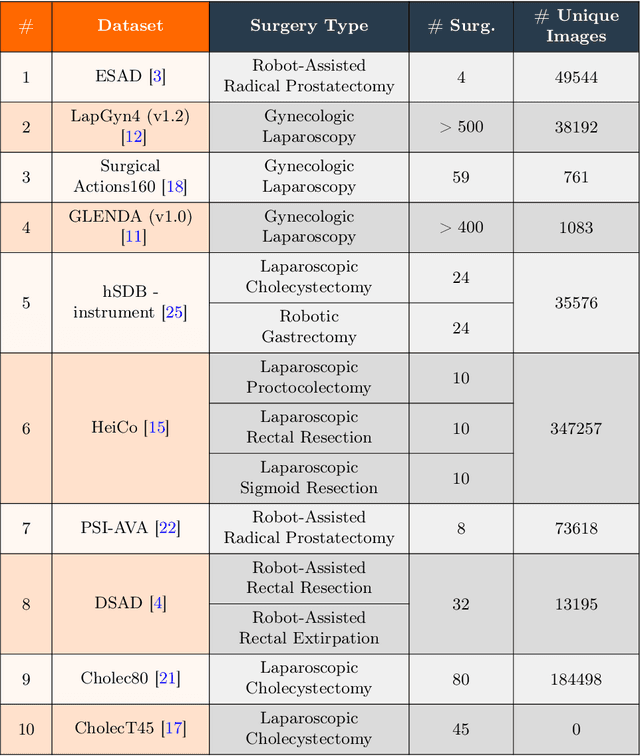
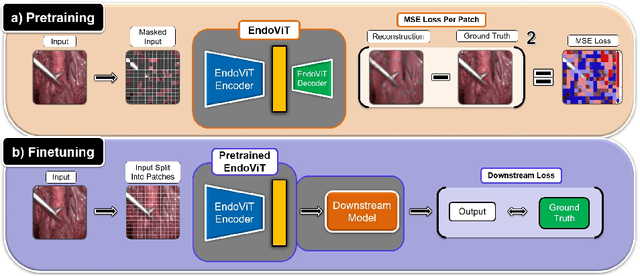
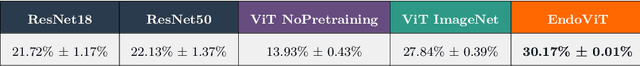
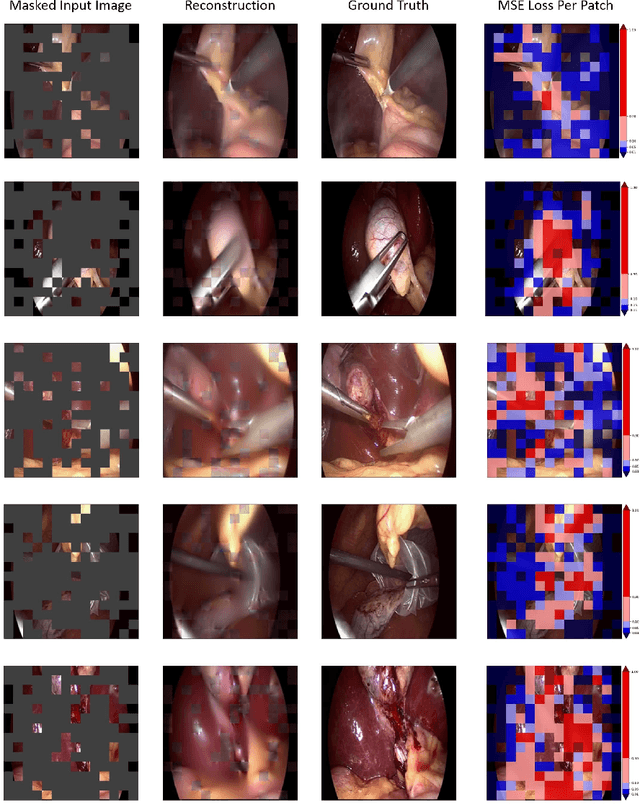
Abstract:Automated endoscopy video analysis is a challenging task in medical computer vision, with the primary objective of assisting surgeons during procedures. The difficulty arises from the complexity of surgical scenes and the lack of a sufficient amount of annotated data. In recent years, large-scale pretraining has shown great success in natural language processing and computer vision communities. These approaches reduce the need for annotated data, which is always a concern in the medical domain. However, most works on endoscopic video understanding use models pretrained on natural images, creating a domain gap between pretraining and finetuning. In this work, we investigate the need for endoscopy domain-specific pretraining based on downstream objectives. To this end, we first collect Endo700k, the largest publicly available corpus of endoscopic images, extracted from nine public Minimally Invasive Surgery (MIS) datasets. Endo700k comprises more than 700,000 unannotated raw images. Next, we introduce EndoViT, an endoscopy pretrained Vision Transformer (ViT). Through ablations, we demonstrate that domain-specific pretraining is particularly beneficial for more complex downstream tasks, such as Action Triplet Detection, and less effective and even unnecessary for simpler tasks, such as Surgical Phase Recognition. We will release both our code and pretrained models upon acceptance to facilitate further research in this direction.
LABRAD-OR: Lightweight Memory Scene Graphs for Accurate Bimodal Reasoning in Dynamic Operating Rooms
Mar 23, 2023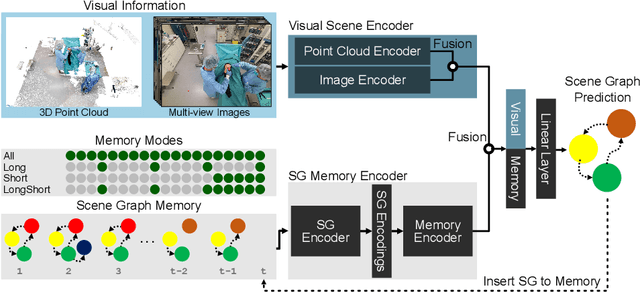

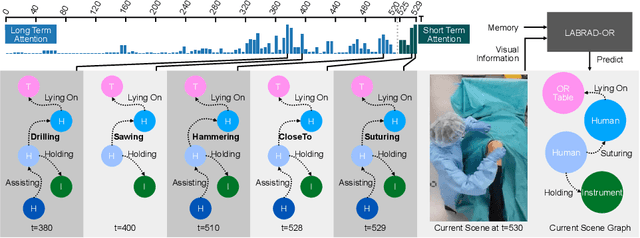

Abstract:Modern surgeries are performed in complex and dynamic settings, including ever-changing interactions between medical staff, patients, and equipment. The holistic modeling of the operating room (OR) is, therefore, a challenging but essential task, with the potential to optimize the performance of surgical teams and aid in developing new surgical technologies to improve patient outcomes. The holistic representation of surgical scenes as semantic scene graphs (SGG), where entities are represented as nodes and relations between them as edges, is a promising direction for fine-grained semantic OR understanding. We propose, for the first time, the use of temporal information for more accurate and consistent holistic OR modeling. Specifically, we introduce memory scene graphs, where the scene graphs of previous time steps act as the temporal representation guiding the current prediction. We design an end-to-end architecture that intelligently fuses the temporal information of our lightweight memory scene graphs with the visual information from point clouds and images. We evaluate our method on the 4D-OR dataset and demonstrate that integrating temporality leads to more accurate and consistent results achieving an +5% increase and a new SOTA of 0.88 in macro F1. This work opens the path for representing the entire surgery history with memory scene graphs and improves the holistic understanding in the OR. Introducing scene graphs as memory representations can offer a valuable tool for many temporal understanding tasks.
Location-Free Scene Graph Generation
Mar 20, 2023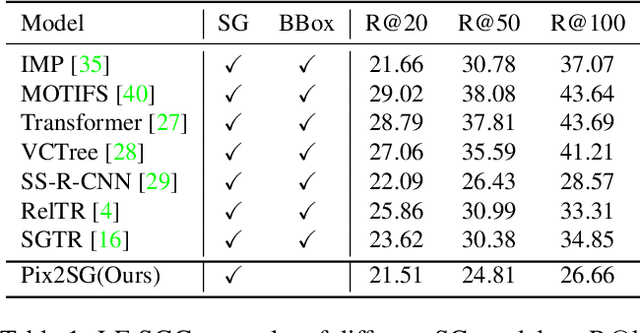
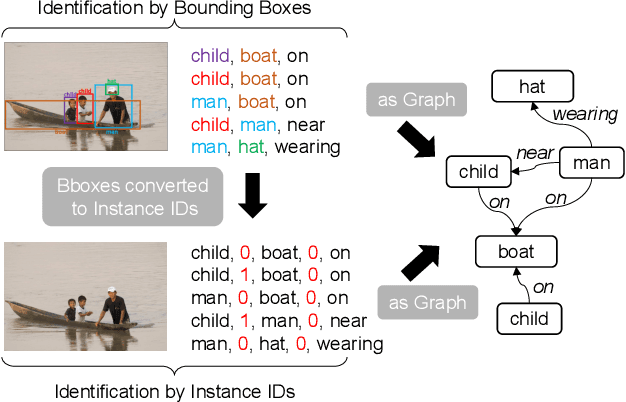
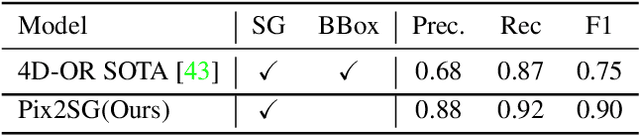
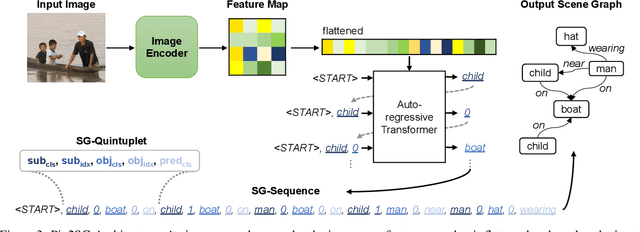
Abstract:Scene Graph Generation (SGG) is a challenging visual understanding task. It combines the detection of entities and relationships between them in a scene. Both previous works and existing evaluation metrics rely on bounding box labels, even though many downstream scene graph applications do not need location information. The need for localization labels significantly increases the annotation cost and hampers the creation of more and larger scene graph datasets. We suggest breaking the dependency of scene graphs on bounding box labels by proposing location-free scene graph generation (LF-SGG). This new task aims at predicting instances of entities, as well as their relationships, without spatial localization. To objectively evaluate the task, the predicted and ground truth scene graphs need to be compared. We solve this NP-hard problem through an efficient algorithm using branching. Additionally, we design the first LF-SGG method, Pix2SG, using autoregressive sequence modeling. Our proposed method is evaluated on Visual Genome and 4D-OR. Although using significantly fewer labels during training, we achieve 74.12\% of the location-supervised SOTA performance on Visual Genome and even outperform the best method on 4D-OR.
CholecTriplet2022: Show me a tool and tell me the triplet -- an endoscopic vision challenge for surgical action triplet detection
Feb 13, 2023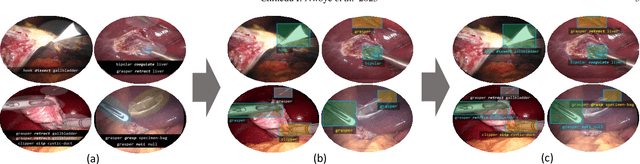
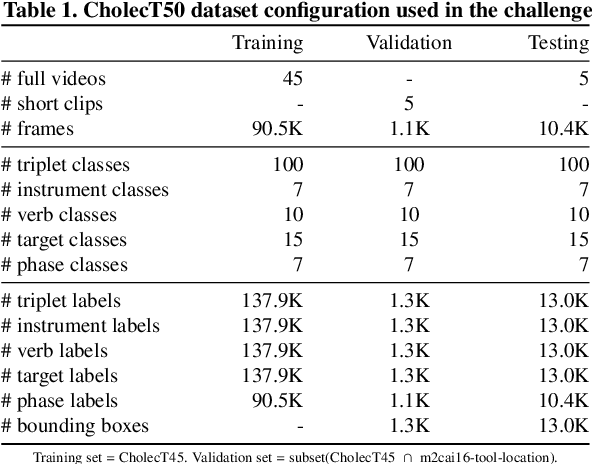
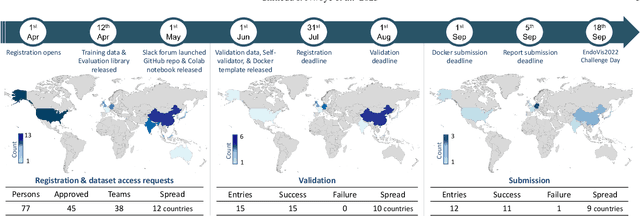
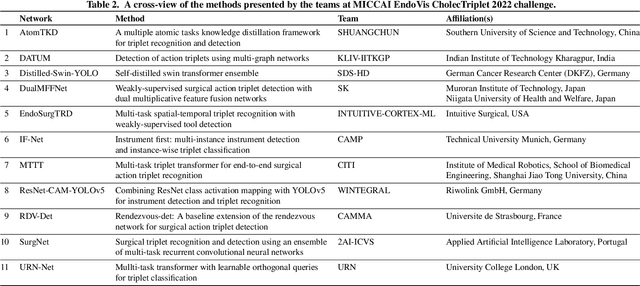
Abstract:Formalizing surgical activities as triplets of the used instruments, actions performed, and target anatomies is becoming a gold standard approach for surgical activity modeling. The benefit is that this formalization helps to obtain a more detailed understanding of tool-tissue interaction which can be used to develop better Artificial Intelligence assistance for image-guided surgery. Earlier efforts and the CholecTriplet challenge introduced in 2021 have put together techniques aimed at recognizing these triplets from surgical footage. Estimating also the spatial locations of the triplets would offer a more precise intraoperative context-aware decision support for computer-assisted intervention. This paper presents the CholecTriplet2022 challenge, which extends surgical action triplet modeling from recognition to detection. It includes weakly-supervised bounding box localization of every visible surgical instrument (or tool), as the key actors, and the modeling of each tool-activity in the form of <instrument, verb, target> triplet. The paper describes a baseline method and 10 new deep learning algorithms presented at the challenge to solve the task. It also provides thorough methodological comparisons of the methods, an in-depth analysis of the obtained results, their significance, and useful insights for future research directions and applications in surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge