Tobias Czempiel
Mitigating Biases in Surgical Operating Rooms with Geometry
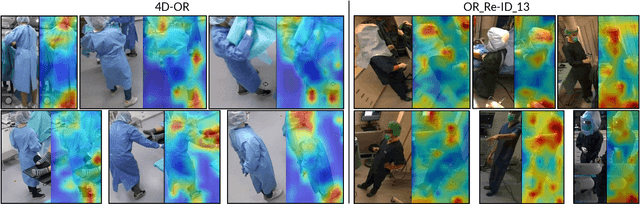
Aug 11, 2025Abstract:Deep neural networks are prone to learning spurious correlations, exploiting dataset-specific artifacts rather than meaningful features for prediction. In surgical operating rooms (OR), these manifest through the standardization of smocks and gowns that obscure robust identifying landmarks, introducing model bias for tasks related to modeling OR personnel. Through gradient-based saliency analysis on two public OR datasets, we reveal that CNN models succumb to such shortcuts, fixating on incidental visual cues such as footwear beneath surgical gowns, distinctive eyewear, or other role-specific identifiers. Avoiding such biases is essential for the next generation of intelligent assistance systems in the OR, which should accurately recognize personalized workflow traits, such as surgical skill level or coordination with other staff members. We address this problem by encoding personnel as 3D point cloud sequences, disentangling identity-relevant shape and motion patterns from appearance-based confounders. Our experiments demonstrate that while RGB and geometric methods achieve comparable performance on datasets with apparent simulation artifacts, RGB models suffer a 12% accuracy drop in realistic clinical settings with decreased visual diversity due to standardizations. This performance gap confirms that geometric representations capture more meaningful biometric features, providing an avenue to developing robust methods of modeling humans in the OR.
SAMSA: Segment Anything Model Enhanced with Spectral Angles for Hyperspectral Interactive Medical Image Segmentation
Jul 31, 2025Abstract:Hyperspectral imaging (HSI) provides rich spectral information for medical imaging, yet encounters significant challenges due to data limitations and hardware variations. We introduce SAMSA, a novel interactive segmentation framework that combines an RGB foundation model with spectral analysis. SAMSA efficiently utilizes user clicks to guide both RGB segmentation and spectral similarity computations. The method addresses key limitations in HSI segmentation through a unique spectral feature fusion strategy that operates independently of spectral band count and resolution. Performance evaluation on publicly available datasets has shown 81.0% 1-click and 93.4% 5-click DICE on a neurosurgical and 81.1% 1-click and 89.2% 5-click DICE on an intraoperative porcine hyperspectral dataset. Experimental results demonstrate SAMSA's effectiveness in few-shot and zero-shot learning scenarios and using minimal training examples. Our approach enables seamless integration of datasets with different spectral characteristics, providing a flexible framework for hyperspectral medical image analysis.
Beyond Role-Based Surgical Domain Modeling: Generalizable Re-Identification in the Operating Room
Mar 17, 2025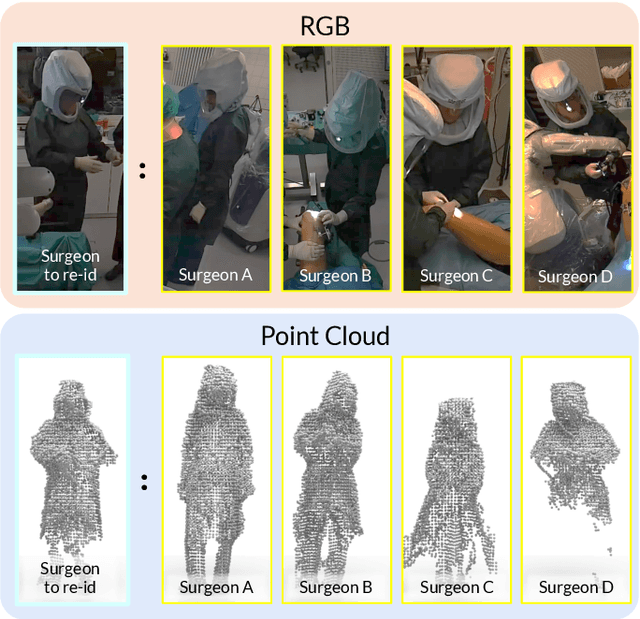

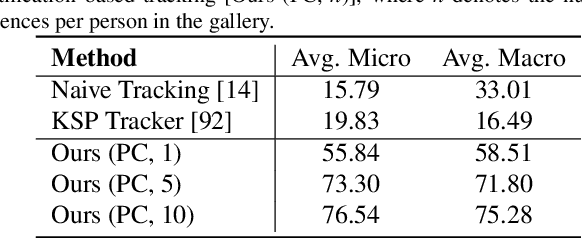
Abstract:Surgical domain models improve workflow optimization through automated predictions of each staff member's surgical role. However, mounting evidence indicates that team familiarity and individuality impact surgical outcomes. We present a novel staff-centric modeling approach that characterizes individual team members through their distinctive movement patterns and physical characteristics, enabling long-term tracking and analysis of surgical personnel across multiple procedures. To address the challenge of inter-clinic variability, we develop a generalizable re-identification framework that encodes sequences of 3D point clouds to capture shape and articulated motion patterns unique to each individual. Our method achieves 86.19% accuracy on realistic clinical data while maintaining 75.27% accuracy when transferring between different environments - a 12% improvement over existing methods. When used to augment markerless personnel tracking, our approach improves accuracy by over 50%. Through extensive validation across three datasets and the introduction of a novel workflow visualization technique, we demonstrate how our framework can reveal novel insights into surgical team dynamics and space utilization patterns, advancing methods to analyze surgical workflows and team coordination.
MM-OR: A Large Multimodal Operating Room Dataset for Semantic Understanding of High-Intensity Surgical Environments
Mar 04, 2025
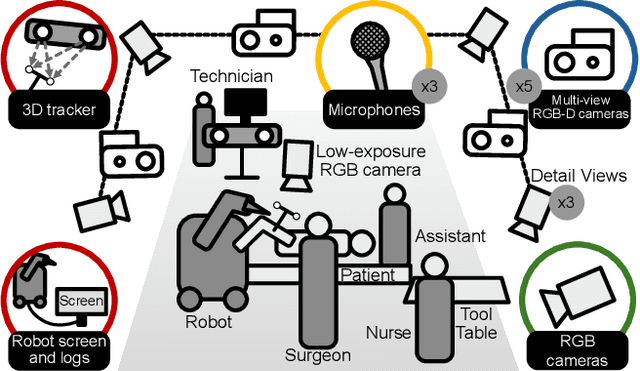
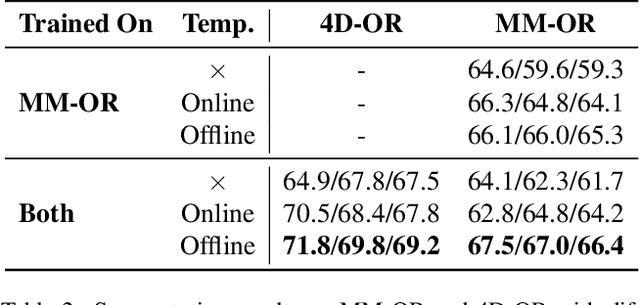
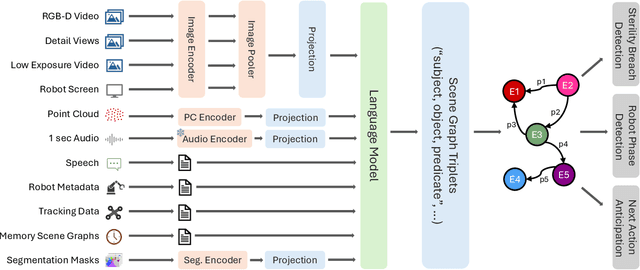
Abstract:Operating rooms (ORs) are complex, high-stakes environments requiring precise understanding of interactions among medical staff, tools, and equipment for enhancing surgical assistance, situational awareness, and patient safety. Current datasets fall short in scale, realism and do not capture the multimodal nature of OR scenes, limiting progress in OR modeling. To this end, we introduce MM-OR, a realistic and large-scale multimodal spatiotemporal OR dataset, and the first dataset to enable multimodal scene graph generation. MM-OR captures comprehensive OR scenes containing RGB-D data, detail views, audio, speech transcripts, robotic logs, and tracking data and is annotated with panoptic segmentations, semantic scene graphs, and downstream task labels. Further, we propose MM2SG, the first multimodal large vision-language model for scene graph generation, and through extensive experiments, demonstrate its ability to effectively leverage multimodal inputs. Together, MM-OR and MM2SG establish a new benchmark for holistic OR understanding, and open the path towards multimodal scene analysis in complex, high-stakes environments. Our code, and data is available at https://github.com/egeozsoy/MM-OR.
RGB to Hyperspectral: Spectral Reconstruction for Enhanced Surgical Imaging
Oct 17, 2024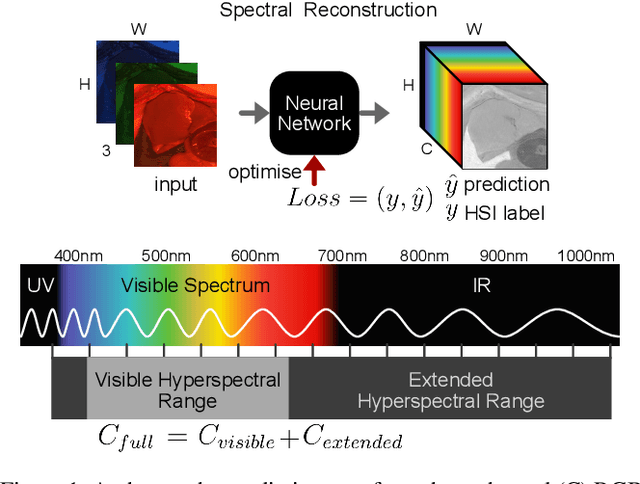



Abstract:This study investigates the reconstruction of hyperspectral signatures from RGB data to enhance surgical imaging, utilizing the publicly available HeiPorSPECTRAL dataset from porcine surgery and an in-house neurosurgery dataset. Various architectures based on convolutional neural networks (CNNs) and transformer models are evaluated using comprehensive metrics. Transformer models exhibit superior performance in terms of RMSE, SAM, PSNR and SSIM by effectively integrating spatial information to predict accurate spectral profiles, encompassing both visible and extended spectral ranges. Qualitative assessments demonstrate the capability to predict spectral profiles critical for informed surgical decision-making during procedures. Challenges associated with capturing both the visible and extended hyperspectral ranges are highlighted using the MAE, emphasizing the complexities involved. The findings open up the new research direction of hyperspectral reconstruction for surgical applications and clinical use cases in real-time surgical environments.
Robotic Arm Platform for Multi-View Image Acquisition and 3D Reconstruction in Minimally Invasive Surgery
Oct 15, 2024Abstract:Minimally invasive surgery (MIS) offers significant benefits such as reduced recovery time and minimised patient trauma, but poses challenges in visibility and access, making accurate 3D reconstruction a significant tool in surgical planning and navigation. This work introduces a robotic arm platform for efficient multi-view image acquisition and precise 3D reconstruction in MIS settings. We adapted a laparoscope to a robotic arm and captured ex-vivo images of several ovine organs across varying lighting conditions (operating room and laparoscopic) and trajectories (spherical and laparoscopic). We employed recently released learning-based feature matchers combined with COLMAP to produce our reconstructions. The reconstructions were evaluated against high-precision laser scans for quantitative evaluation. Our results show that whilst reconstructions suffer most under realistic MIS lighting and trajectory, many versions of our pipeline achieve close to sub-millimetre accuracy with an average of 1.05 mm Root Mean Squared Error and 0.82 mm Chamfer distance. Our best reconstruction results occur with operating room lighting and spherical trajectories. Our robotic platform provides a tool for controlled, repeatable multi-view data acquisition for 3D generation in MIS environments which we hope leads to new datasets for training learning-based models.
Dynamic Scene Graph Representation for Surgical Video
Sep 25, 2023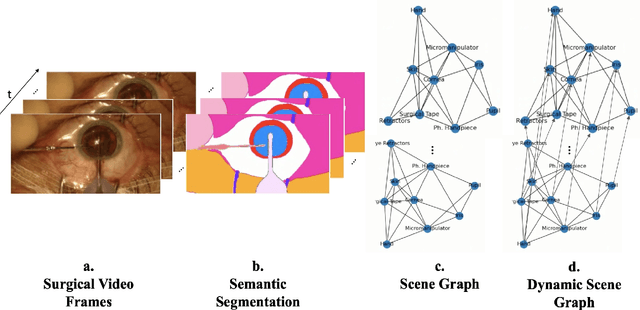
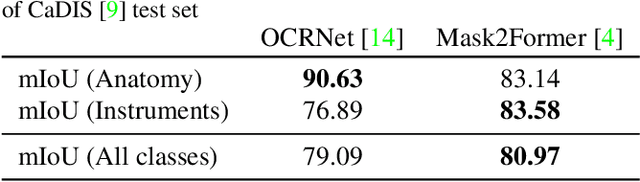
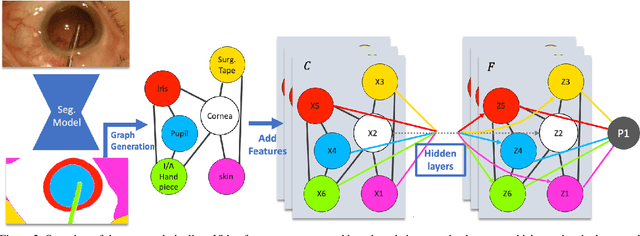
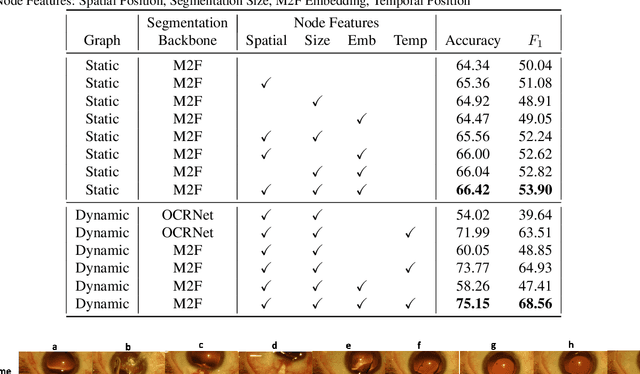
Abstract:Surgical videos captured from microscopic or endoscopic imaging devices are rich but complex sources of information, depicting different tools and anatomical structures utilized during an extended amount of time. Despite containing crucial workflow information and being commonly recorded in many procedures, usage of surgical videos for automated surgical workflow understanding is still limited. In this work, we exploit scene graphs as a more holistic, semantically meaningful and human-readable way to represent surgical videos while encoding all anatomical structures, tools, and their interactions. To properly evaluate the impact of our solutions, we create a scene graph dataset from semantic segmentations from the CaDIS and CATARACTS datasets. We demonstrate that scene graphs can be leveraged through the use of graph convolutional networks (GCNs) to tackle surgical downstream tasks such as surgical workflow recognition with competitive performance. Moreover, we demonstrate the benefits of surgical scene graphs regarding the explainability and robustness of model decisions, which are crucial in the clinical setting.
DisguisOR: Holistic Face Anonymization for the Operating Room
Jul 26, 2023Abstract:Purpose: Recent advances in Surgical Data Science (SDS) have contributed to an increase in video recordings from hospital environments. While methods such as surgical workflow recognition show potential in increasing the quality of patient care, the quantity of video data has surpassed the scale at which images can be manually anonymized. Existing automated 2D anonymization methods under-perform in Operating Rooms (OR), due to occlusions and obstructions. We propose to anonymize multi-view OR recordings using 3D data from multiple camera streams. Methods: RGB and depth images from multiple cameras are fused into a 3D point cloud representation of the scene. We then detect each individual's face in 3D by regressing a parametric human mesh model onto detected 3D human keypoints and aligning the face mesh with the fused 3D point cloud. The mesh model is rendered into every acquired camera view, replacing each individual's face. Results: Our method shows promise in locating faces at a higher rate than existing approaches. DisguisOR produces geometrically consistent anonymizations for each camera view, enabling more realistic anonymization that is less detrimental to downstream tasks. Conclusion: Frequent obstructions and crowding in operating rooms leaves significant room for improvement for off-the-shelf anonymization methods. DisguisOR addresses privacy on a scene level and has the potential to facilitate further research in SDS.
Whether and When does Endoscopy Domain Pretraining Make Sense?
Mar 30, 2023
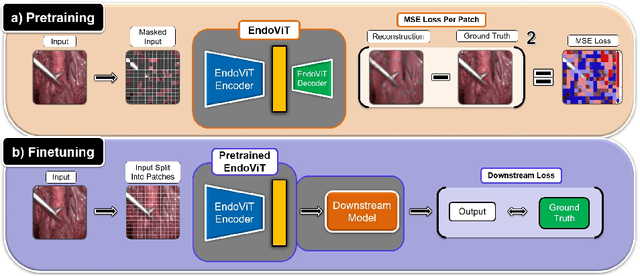
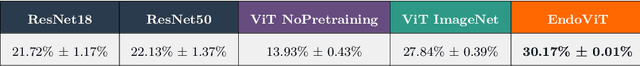
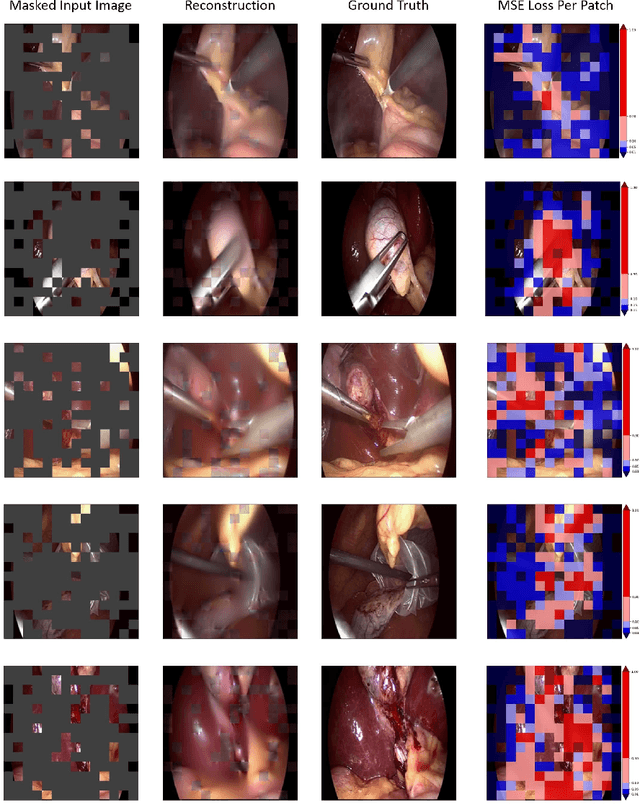
Abstract:Automated endoscopy video analysis is a challenging task in medical computer vision, with the primary objective of assisting surgeons during procedures. The difficulty arises from the complexity of surgical scenes and the lack of a sufficient amount of annotated data. In recent years, large-scale pretraining has shown great success in natural language processing and computer vision communities. These approaches reduce the need for annotated data, which is always a concern in the medical domain. However, most works on endoscopic video understanding use models pretrained on natural images, creating a domain gap between pretraining and finetuning. In this work, we investigate the need for endoscopy domain-specific pretraining based on downstream objectives. To this end, we first collect Endo700k, the largest publicly available corpus of endoscopic images, extracted from nine public Minimally Invasive Surgery (MIS) datasets. Endo700k comprises more than 700,000 unannotated raw images. Next, we introduce EndoViT, an endoscopy pretrained Vision Transformer (ViT). Through ablations, we demonstrate that domain-specific pretraining is particularly beneficial for more complex downstream tasks, such as Action Triplet Detection, and less effective and even unnecessary for simpler tasks, such as Surgical Phase Recognition. We will release both our code and pretrained models upon acceptance to facilitate further research in this direction.
LABRAD-OR: Lightweight Memory Scene Graphs for Accurate Bimodal Reasoning in Dynamic Operating Rooms
Mar 23, 2023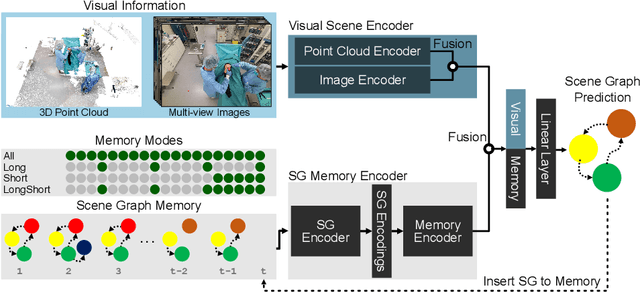

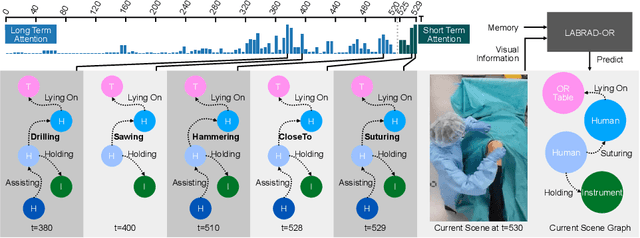

Abstract:Modern surgeries are performed in complex and dynamic settings, including ever-changing interactions between medical staff, patients, and equipment. The holistic modeling of the operating room (OR) is, therefore, a challenging but essential task, with the potential to optimize the performance of surgical teams and aid in developing new surgical technologies to improve patient outcomes. The holistic representation of surgical scenes as semantic scene graphs (SGG), where entities are represented as nodes and relations between them as edges, is a promising direction for fine-grained semantic OR understanding. We propose, for the first time, the use of temporal information for more accurate and consistent holistic OR modeling. Specifically, we introduce memory scene graphs, where the scene graphs of previous time steps act as the temporal representation guiding the current prediction. We design an end-to-end architecture that intelligently fuses the temporal information of our lightweight memory scene graphs with the visual information from point clouds and images. We evaluate our method on the 4D-OR dataset and demonstrate that integrating temporality leads to more accurate and consistent results achieving an +5% increase and a new SOTA of 0.88 in macro F1. This work opens the path for representing the entire surgery history with memory scene graphs and improves the holistic understanding in the OR. Introducing scene graphs as memory representations can offer a valuable tool for many temporal understanding tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge