Matthias Keicher
Language Agents for Hypothesis-driven Clinical Decision Making with Reinforcement Learning
Jun 16, 2025Abstract:Clinical decision-making is a dynamic, interactive, and cyclic process where doctors have to repeatedly decide on which clinical action to perform and consider newly uncovered information for diagnosis and treatment. Large Language Models (LLMs) have the potential to support clinicians in this process, however, most applications of LLMs in clinical decision support suffer from one of two limitations: Either they assume the unrealistic scenario of immediate availability of all patient information and do not model the interactive and iterative investigation process, or they restrict themselves to the limited "out-of-the-box" capabilities of large pre-trained models without performing task-specific training. In contrast to this, we propose to model clinical decision-making for diagnosis with a hypothesis-driven uncertainty-aware language agent, LA-CDM, that converges towards a diagnosis via repeatedly requesting and interpreting relevant tests. Using a hybrid training paradigm combining supervised and reinforcement learning, we train LA-CDM with three objectives targeting critical aspects of clinical decision-making: accurate hypothesis generation, hypothesis uncertainty estimation, and efficient decision-making. We evaluate our methodology on MIMIC-CDM, a real-world dataset covering four abdominal diseases containing various clinical tests and show the benefit of explicitly training clinical decision-making for increasing diagnostic performance and efficiency.
From EHRs to Patient Pathways: Scalable Modeling of Longitudinal Health Trajectories with LLMs
Jun 05, 2025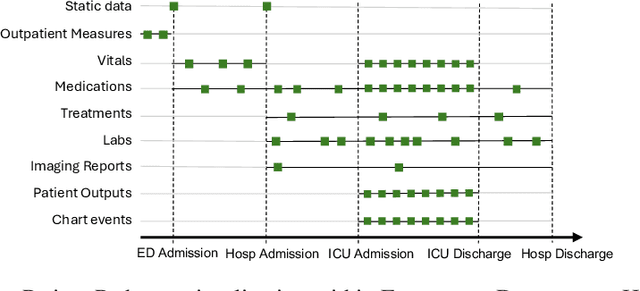
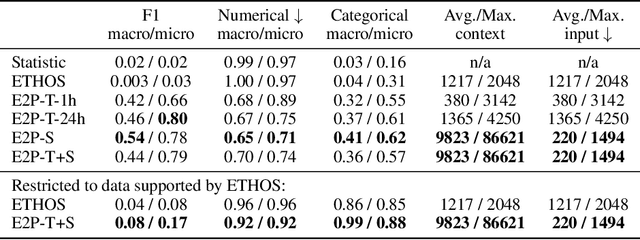
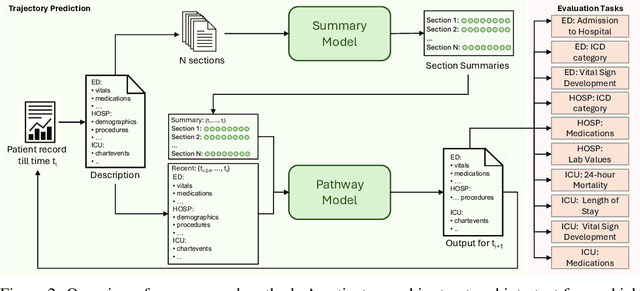
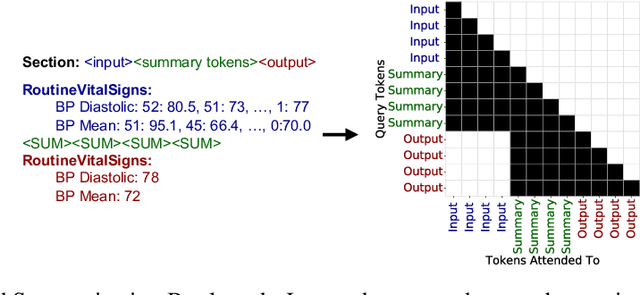
Abstract:Healthcare systems face significant challenges in managing and interpreting vast, heterogeneous patient data for personalized care. Existing approaches often focus on narrow use cases with a limited feature space, overlooking the complex, longitudinal interactions needed for a holistic understanding of patient health. In this work, we propose a novel approach to patient pathway modeling by transforming diverse electronic health record (EHR) data into a structured representation and designing a holistic pathway prediction model, EHR2Path, optimized to predict future health trajectories. Further, we introduce a novel summary mechanism that embeds long-term temporal context into topic-specific summary tokens, improving performance over text-only models, while being much more token-efficient. EHR2Path demonstrates strong performance in both next time-step prediction and longitudinal simulation, outperforming competitive baselines. It enables detailed simulations of patient trajectories, inherently targeting diverse evaluation tasks, such as forecasting vital signs, lab test results, or length-of-stay, opening a path towards predictive and personalized healthcare.
ORQA: A Benchmark and Foundation Model for Holistic Operating Room Modeling
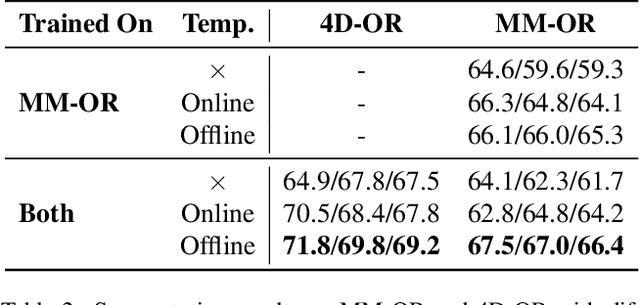
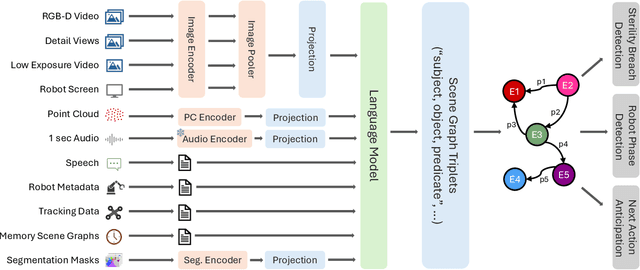
May 19, 2025Abstract:The real-world complexity of surgeries necessitates surgeons to have deep and holistic comprehension to ensure precision, safety, and effective interventions. Computational systems are required to have a similar level of comprehension within the operating room. Prior works, limited to single-task efforts like phase recognition or scene graph generation, lack scope and generalizability. In this work, we introduce ORQA, a novel OR question answering benchmark and foundational multimodal model to advance OR intelligence. By unifying all four public OR datasets into a comprehensive benchmark, we enable our approach to concurrently address a diverse range of OR challenges. The proposed multimodal large language model fuses diverse OR signals such as visual, auditory, and structured data, for a holistic modeling of the OR. Finally, we propose a novel, progressive knowledge distillation paradigm, to generate a family of models optimized for different speed and memory requirements. We show the strong performance of ORQA on our proposed benchmark, and its zero-shot generalization, paving the way for scalable, unified OR modeling and significantly advancing multimodal surgical intelligence. We will release our code and data upon acceptance.
Rewarding Doubt: A Reinforcement Learning Approach to Confidence Calibration of Large Language Models
Mar 05, 2025Abstract:A safe and trustworthy use of Large Language Models (LLMs) requires an accurate expression of confidence in their answers. We introduce a novel Reinforcement Learning (RL) approach for LLM calibration that fine-tunes LLMs to elicit calibrated confidence estimations in their answers to factual questions. We model the problem as a betting game where the model predicts a confidence score together with every answer, and design a reward function that penalizes both over and under-confidence. We prove that under our reward design an optimal policy would result in a perfectly calibrated confidence estimation. Our experiments demonstrate significantly improved confidence calibration and generalization to new tasks without re-training, indicating that our approach teaches a general confidence awareness. This approach enables the training of inherently calibrated LLMs.
MM-OR: A Large Multimodal Operating Room Dataset for Semantic Understanding of High-Intensity Surgical Environments
Mar 04, 2025
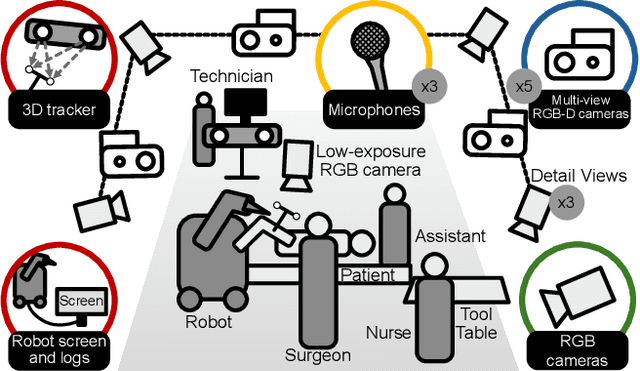
Abstract:Operating rooms (ORs) are complex, high-stakes environments requiring precise understanding of interactions among medical staff, tools, and equipment for enhancing surgical assistance, situational awareness, and patient safety. Current datasets fall short in scale, realism and do not capture the multimodal nature of OR scenes, limiting progress in OR modeling. To this end, we introduce MM-OR, a realistic and large-scale multimodal spatiotemporal OR dataset, and the first dataset to enable multimodal scene graph generation. MM-OR captures comprehensive OR scenes containing RGB-D data, detail views, audio, speech transcripts, robotic logs, and tracking data and is annotated with panoptic segmentations, semantic scene graphs, and downstream task labels. Further, we propose MM2SG, the first multimodal large vision-language model for scene graph generation, and through extensive experiments, demonstrate its ability to effectively leverage multimodal inputs. Together, MM-OR and MM2SG establish a new benchmark for holistic OR understanding, and open the path towards multimodal scene analysis in complex, high-stakes environments. Our code, and data is available at https://github.com/egeozsoy/MM-OR.
From large language models to multimodal AI: A scoping review on the potential of generative AI in medicine
Feb 13, 2025



Abstract:Generative artificial intelligence (AI) models, such as diffusion models and OpenAI's ChatGPT, are transforming medicine by enhancing diagnostic accuracy and automating clinical workflows. The field has advanced rapidly, evolving from text-only large language models for tasks such as clinical documentation and decision support to multimodal AI systems capable of integrating diverse data modalities, including imaging, text, and structured data, within a single model. The diverse landscape of these technologies, along with rising interest, highlights the need for a comprehensive review of their applications and potential. This scoping review explores the evolution of multimodal AI, highlighting its methods, applications, datasets, and evaluation in clinical settings. Adhering to PRISMA-ScR guidelines, we systematically queried PubMed, IEEE Xplore, and Web of Science, prioritizing recent studies published up to the end of 2024. After rigorous screening, 144 papers were included, revealing key trends and challenges in this dynamic field. Our findings underscore a shift from unimodal to multimodal approaches, driving innovations in diagnostic support, medical report generation, drug discovery, and conversational AI. However, critical challenges remain, including the integration of heterogeneous data types, improving model interpretability, addressing ethical concerns, and validating AI systems in real-world clinical settings. This review summarizes the current state of the art, identifies critical gaps, and provides insights to guide the development of scalable, trustworthy, and clinically impactful multimodal AI solutions in healthcare.
MAGDA: Multi-agent guideline-driven diagnostic assistance
Sep 10, 2024Abstract:In emergency departments, rural hospitals, or clinics in less developed regions, clinicians often lack fast image analysis by trained radiologists, which can have a detrimental effect on patients' healthcare. Large Language Models (LLMs) have the potential to alleviate some pressure from these clinicians by providing insights that can help them in their decision-making. While these LLMs achieve high test results on medical exams showcasing their great theoretical medical knowledge, they tend not to follow medical guidelines. In this work, we introduce a new approach for zero-shot guideline-driven decision support. We model a system of multiple LLM agents augmented with a contrastive vision-language model that collaborate to reach a patient diagnosis. After providing the agents with simple diagnostic guidelines, they will synthesize prompts and screen the image for findings following these guidelines. Finally, they provide understandable chain-of-thought reasoning for their diagnosis, which is then self-refined to consider inter-dependencies between diseases. As our method is zero-shot, it is adaptable to settings with rare diseases, where training data is limited, but expert-crafted disease descriptions are available. We evaluate our method on two chest X-ray datasets, CheXpert and ChestX-ray 14 Longtail, showcasing performance improvement over existing zero-shot methods and generalizability to rare diseases.
Counterfactual Explanations for Medical Image Classification and Regression using Diffusion Autoencoder
Aug 02, 2024



Abstract:Counterfactual explanations (CEs) aim to enhance the interpretability of machine learning models by illustrating how alterations in input features would affect the resulting predictions. Common CE approaches require an additional model and are typically constrained to binary counterfactuals. In contrast, we propose a novel method that operates directly on the latent space of a generative model, specifically a Diffusion Autoencoder (DAE). This approach offers inherent interpretability by enabling the generation of CEs and the continuous visualization of the model's internal representation across decision boundaries. Our method leverages the DAE's ability to encode images into a semantically rich latent space in an unsupervised manner, eliminating the need for labeled data or separate feature extraction models. We show that these latent representations are helpful for medical condition classification and the ordinal regression of severity pathologies, such as vertebral compression fractures (VCF) and diabetic retinopathy (DR). Beyond binary CEs, our method supports the visualization of ordinal CEs using a linear model, providing deeper insights into the model's decision-making process and enhancing interpretability. Experiments across various medical imaging datasets demonstrate the method's advantages in interpretability and versatility. The linear manifold of the DAE's latent space allows for meaningful interpolation and manipulation, making it a powerful tool for exploring medical image properties. Our code is available at https://github.com/matanat/dae_counterfactual.
ORacle: Large Vision-Language Models for Knowledge-Guided Holistic OR Domain Modeling
Apr 10, 2024



Abstract:Every day, countless surgeries are performed worldwide, each within the distinct settings of operating rooms (ORs) that vary not only in their setups but also in the personnel, tools, and equipment used. This inherent diversity poses a substantial challenge for achieving a holistic understanding of the OR, as it requires models to generalize beyond their initial training datasets. To reduce this gap, we introduce ORacle, an advanced vision-language model designed for holistic OR domain modeling, which incorporates multi-view and temporal capabilities and can leverage external knowledge during inference, enabling it to adapt to previously unseen surgical scenarios. This capability is further enhanced by our novel data augmentation framework, which significantly diversifies the training dataset, ensuring ORacle's proficiency in applying the provided knowledge effectively. In rigorous testing, in scene graph generation, and downstream tasks on the 4D-OR dataset, ORacle not only demonstrates state-of-the-art performance but does so requiring less data than existing models. Furthermore, its adaptability is displayed through its ability to interpret unseen views, actions, and appearances of tools and equipment. This demonstrates ORacle's potential to significantly enhance the scalability and affordability of OR domain modeling and opens a pathway for future advancements in surgical data science. We will release our code and data upon acceptance.
RaDialog: A Large Vision-Language Model for Radiology Report Generation and Conversational Assistance
Nov 30, 2023



Abstract:Conversational AI tools that can generate and discuss clinically correct radiology reports for a given medical image have the potential to transform radiology. Such a human-in-the-loop radiology assistant could facilitate a collaborative diagnostic process, thus saving time and improving the quality of reports. Towards this goal, we introduce RaDialog, the first thoroughly evaluated and publicly available large vision-language model for radiology report generation and interactive dialog. RaDialog effectively integrates visual image features and structured pathology findings with a large language model (LLM) while simultaneously adapting it to a specialized domain using parameter-efficient fine-tuning. To keep the conversational abilities of the underlying LLM, we propose a comprehensive, semi-automatically labeled, image-grounded instruct dataset for chest X-ray radiology tasks. By training with this dataset, our method achieves state-of-the-art clinical correctness in report generation and shows impressive abilities in interactive tasks such as correcting reports and answering questions, serving as a foundational step toward clinical dialog systems. Our code is available on github: https://github.com/ChantalMP/RaDialog.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge