Sophia Bano
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
Federated Learning for Surgical Vision in Appendicitis Classification: Results of the FedSurg EndoVis 2024 Challenge
Oct 06, 2025Abstract:Purpose: The FedSurg challenge was designed to benchmark the state of the art in federated learning for surgical video classification. Its goal was to assess how well current methods generalize to unseen clinical centers and adapt through local fine-tuning while enabling collaborative model development without sharing patient data. Methods: Participants developed strategies to classify inflammation stages in appendicitis using a preliminary version of the multi-center Appendix300 video dataset. The challenge evaluated two tasks: generalization to an unseen center and center-specific adaptation after fine-tuning. Submitted approaches included foundation models with linear probing, metric learning with triplet loss, and various FL aggregation schemes (FedAvg, FedMedian, FedSAM). Performance was assessed using F1-score and Expected Cost, with ranking robustness evaluated via bootstrapping and statistical testing. Results: In the generalization task, performance across centers was limited. In the adaptation task, all teams improved after fine-tuning, though ranking stability was low. The ViViT-based submission achieved the strongest overall performance. The challenge highlighted limitations in generalization, sensitivity to class imbalance, and difficulties in hyperparameter tuning in decentralized training, while spatiotemporal modeling and context-aware preprocessing emerged as promising strategies. Conclusion: The FedSurg Challenge establishes the first benchmark for evaluating FL strategies in surgical video classification. Findings highlight the trade-off between local personalization and global robustness, and underscore the importance of architecture choice, preprocessing, and loss design. This benchmarking offers a reference point for future development of imbalance-aware, adaptive, and robust FL methods in clinical surgical AI.
Robotic Arm Platform for Multi-View Image Acquisition and 3D Reconstruction in Minimally Invasive Surgery
Oct 15, 2024Abstract:Minimally invasive surgery (MIS) offers significant benefits such as reduced recovery time and minimised patient trauma, but poses challenges in visibility and access, making accurate 3D reconstruction a significant tool in surgical planning and navigation. This work introduces a robotic arm platform for efficient multi-view image acquisition and precise 3D reconstruction in MIS settings. We adapted a laparoscope to a robotic arm and captured ex-vivo images of several ovine organs across varying lighting conditions (operating room and laparoscopic) and trajectories (spherical and laparoscopic). We employed recently released learning-based feature matchers combined with COLMAP to produce our reconstructions. The reconstructions were evaluated against high-precision laser scans for quantitative evaluation. Our results show that whilst reconstructions suffer most under realistic MIS lighting and trajectory, many versions of our pipeline achieve close to sub-millimetre accuracy with an average of 1.05 mm Root Mean Squared Error and 0.82 mm Chamfer distance. Our best reconstruction results occur with operating room lighting and spherical trajectories. Our robotic platform provides a tool for controlled, repeatable multi-view data acquisition for 3D generation in MIS environments which we hope leads to new datasets for training learning-based models.
Automated Surgical Skill Assessment in Endoscopic Pituitary Surgery using Real-time Instrument Tracking on a High-fidelity Bench-top Phantom
Sep 25, 2024Abstract:Improved surgical skill is generally associated with improved patient outcomes, although assessment is subjective; labour-intensive; and requires domain specific expertise. Automated data driven metrics can alleviate these difficulties, as demonstrated by existing machine learning instrument tracking models in minimally invasive surgery. However, these models have been tested on limited datasets of laparoscopic surgery, with a focus on isolated tasks and robotic surgery. In this paper, a new public dataset is introduced, focusing on simulated surgery, using the nasal phase of endoscopic pituitary surgery as an exemplar. Simulated surgery allows for a realistic yet repeatable environment, meaning the insights gained from automated assessment can be used by novice surgeons to hone their skills on the simulator before moving to real surgery. PRINTNet (Pituitary Real-time INstrument Tracking Network) has been created as a baseline model for this automated assessment. Consisting of DeepLabV3 for classification and segmentation; StrongSORT for tracking; and the NVIDIA Holoscan SDK for real-time performance, PRINTNet achieved 71.9% Multiple Object Tracking Precision running at 22 Frames Per Second. Using this tracking output, a Multilayer Perceptron achieved 87% accuracy in predicting surgical skill level (novice or expert), with the "ratio of total procedure time to instrument visible time" correlated with higher surgical skill. This therefore demonstrates the feasibility of automated surgical skill assessment in simulated endoscopic pituitary surgery. The new publicly available dataset can be found here: https://doi.org/10.5522/04/26511049.
PitRSDNet: Predicting Intra-operative Remaining Surgery Duration in Endoscopic Pituitary Surgery
Sep 25, 2024



Abstract:Accurate intra-operative Remaining Surgery Duration (RSD) predictions allow for anaesthetists to more accurately decide when to administer anaesthetic agents and drugs, as well as to notify hospital staff to send in the next patient. Therefore RSD plays an important role in improving patient care and minimising surgical theatre costs via efficient scheduling. In endoscopic pituitary surgery, it is uniquely challenging due to variable workflow sequences with a selection of optional steps contributing to high variability in surgery duration. This paper presents PitRSDNet for predicting RSD during pituitary surgery, a spatio-temporal neural network model that learns from historical data focusing on workflow sequences. PitRSDNet integrates workflow knowledge into RSD prediction in two forms: 1) multi-task learning for concurrently predicting step and RSD; and 2) incorporating prior steps as context in temporal learning and inference. PitRSDNet is trained and evaluated on a new endoscopic pituitary surgery dataset with 88 videos to show competitive performance improvements over previous statistical and machine learning methods. The findings also highlight how PitRSDNet improve RSD precision on outlier cases utilising the knowledge of prior steps.
PitVis-2023 Challenge: Workflow Recognition in videos of Endoscopic Pituitary Surgery
Sep 02, 2024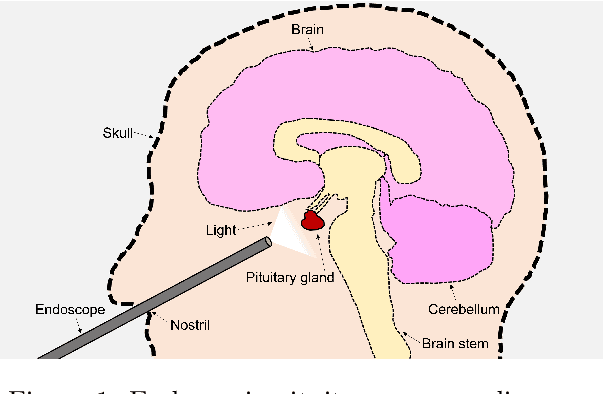
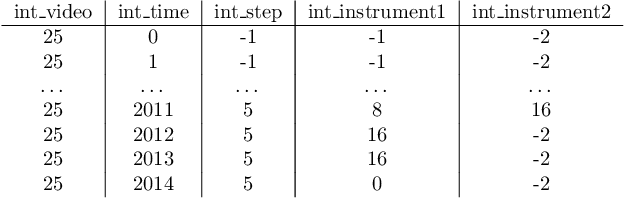
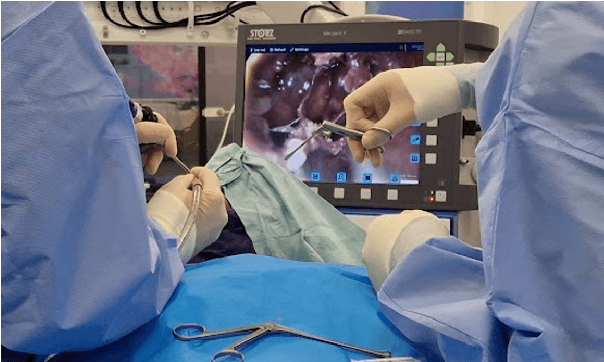
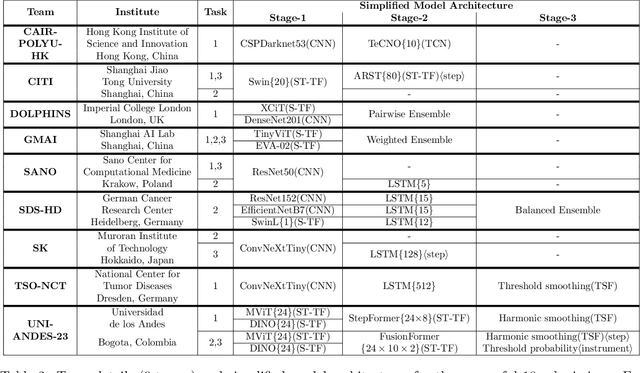
Abstract:The field of computer vision applied to videos of minimally invasive surgery is ever-growing. Workflow recognition pertains to the automated recognition of various aspects of a surgery: including which surgical steps are performed; and which surgical instruments are used. This information can later be used to assist clinicians when learning the surgery; during live surgery; and when writing operation notes. The Pituitary Vision (PitVis) 2023 Challenge tasks the community to step and instrument recognition in videos of endoscopic pituitary surgery. This is a unique task when compared to other minimally invasive surgeries due to the smaller working space, which limits and distorts vision; and higher frequency of instrument and step switching, which requires more precise model predictions. Participants were provided with 25-videos, with results presented at the MICCAI-2023 conference as part of the Endoscopic Vision 2023 Challenge in Vancouver, Canada, on 08-Oct-2023. There were 18-submissions from 9-teams across 6-countries, using a variety of deep learning models. A commonality between the top performing models was incorporating spatio-temporal and multi-task methods, with greater than 50% and 10% macro-F1-score improvement over purely spacial single-task models in step and instrument recognition respectively. The PitVis-2023 Challenge therefore demonstrates state-of-the-art computer vision models in minimally invasive surgery are transferable to a new dataset, with surgery specific techniques used to enhance performance, progressing the field further. Benchmark results are provided in the paper, and the dataset is publicly available at: https://doi.org/10.5522/04/26531686.
DARES: Depth Anything in Robotic Endoscopic Surgery with Self-supervised Vector-LoRA of the Foundation Model
Aug 30, 2024



Abstract:Robotic-assisted surgery (RAS) relies on accurate depth estimation for 3D reconstruction and visualization. While foundation models like Depth Anything Models (DAM) show promise, directly applying them to surgery often yields suboptimal results. Fully fine-tuning on limited surgical data can cause overfitting and catastrophic forgetting, compromising model robustness and generalization. Although Low-Rank Adaptation (LoRA) addresses some adaptation issues, its uniform parameter distribution neglects the inherent feature hierarchy, where earlier layers, learning more general features, require more parameters than later ones. To tackle this issue, we introduce Depth Anything in Robotic Endoscopic Surgery (DARES), a novel approach that employs a new adaptation technique, Vector Low-Rank Adaptation (Vector-LoRA) on the DAM V2 to perform self-supervised monocular depth estimation in RAS scenes. To enhance learning efficiency, we introduce Vector-LoRA by integrating more parameters in earlier layers and gradually decreasing parameters in later layers. We also design a reprojection loss based on the multi-scale SSIM error to enhance depth perception by better tailoring the foundation model to the specific requirements of the surgical environment. The proposed method is validated on the SCARED dataset and demonstrates superior performance over recent state-of-the-art self-supervised monocular depth estimation techniques, achieving an improvement of 13.3% in the absolute relative error metric. The code and pre-trained weights are available at https://github.com/mobarakol/DARES.
Mismatched: Evaluating the Limits of Image Matching Approaches and Benchmarks
Aug 29, 2024



Abstract:Three-dimensional (3D) reconstruction from two-dimensional images is an active research field in computer vision, with applications ranging from navigation and object tracking to segmentation and three-dimensional modeling. Traditionally, parametric techniques have been employed for this task. However, recent advancements have seen a shift towards learning-based methods. Given the rapid pace of research and the frequent introduction of new image matching methods, it is essential to evaluate them. In this paper, we present a comprehensive evaluation of various image matching methods using a structure-from-motion pipeline. We assess the performance of these methods on both in-domain and out-of-domain datasets, identifying key limitations in both the methods and benchmarks. We also investigate the impact of edge detection as a pre-processing step. Our analysis reveals that image matching for 3D reconstruction remains an open challenge, necessitating careful selection and tuning of models for specific scenarios, while also highlighting mismatches in how metrics currently represent method performance.
PitVQA: Image-grounded Text Embedding LLM for Visual Question Answering in Pituitary Surgery
May 22, 2024



Abstract:Visual Question Answering (VQA) within the surgical domain, utilizing Large Language Models (LLMs), offers a distinct opportunity to improve intra-operative decision-making and facilitate intuitive surgeon-AI interaction. However, the development of LLMs for surgical VQA is hindered by the scarcity of diverse and extensive datasets with complex reasoning tasks. Moreover, contextual fusion of the image and text modalities remains an open research challenge due to the inherent differences between these two types of information and the complexity involved in aligning them. This paper introduces PitVQA, a novel dataset specifically designed for VQA in endonasal pituitary surgery and PitVQA-Net, an adaptation of the GPT2 with a novel image-grounded text embedding for surgical VQA. PitVQA comprises 25 procedural videos and a rich collection of question-answer pairs spanning crucial surgical aspects such as phase and step recognition, context understanding, tool detection and localization, and tool-tissue interactions. PitVQA-Net consists of a novel image-grounded text embedding that projects image and text features into a shared embedding space and GPT2 Backbone with an excitation block classification head to generate contextually relevant answers within the complex domain of endonasal pituitary surgery. Our image-grounded text embedding leverages joint embedding, cross-attention and contextual representation to understand the contextual relationship between questions and surgical images. We demonstrate the effectiveness of PitVQA-Net on both the PitVQA and the publicly available EndoVis18-VQA dataset, achieving improvements in balanced accuracy of 8% and 9% over the most recent baselines, respectively. Our code and dataset is available at https://github.com/mobarakol/PitVQA.
Surgical-DeSAM: Decoupling SAM for Instrument Segmentation in Robotic Surgery
Apr 22, 2024Abstract:Purpose: The recent Segment Anything Model (SAM) has demonstrated impressive performance with point, text or bounding box prompts, in various applications. However, in safety-critical surgical tasks, prompting is not possible due to (i) the lack of per-frame prompts for supervised learning, (ii) it is unrealistic to prompt frame-by-frame in a real-time tracking application, and (iii) it is expensive to annotate prompts for offline applications. Methods: We develop Surgical-DeSAM to generate automatic bounding box prompts for decoupling SAM to obtain instrument segmentation in real-time robotic surgery. We utilise a commonly used detection architecture, DETR, and fine-tuned it to obtain bounding box prompt for the instruments. We then empolyed decoupling SAM (DeSAM) by replacing the image encoder with DETR encoder and fine-tune prompt encoder and mask decoder to obtain instance segmentation for the surgical instruments. To improve detection performance, we adopted the Swin-transformer to better feature representation. Results: The proposed method has been validated on two publicly available datasets from the MICCAI surgical instruments segmentation challenge EndoVis 2017 and 2018. The performance of our method is also compared with SOTA instrument segmentation methods and demonstrated significant improvements with dice metrics of 89.62 and 90.70 for the EndoVis 2017 and 2018. Conclusion: Our extensive experiments and validations demonstrate that Surgical-DeSAM enables real-time instrument segmentation without any additional prompting and outperforms other SOTA segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge