Francisco Vasconcelos
A multi-centre, multi-device benchmark dataset for landmark-based comprehensive fetal biometry
Dec 19, 2025Abstract:Accurate fetal growth assessment from ultrasound (US) relies on precise biometry measured by manually identifying anatomical landmarks in standard planes. Manual landmarking is time-consuming, operator-dependent, and sensitive to variability across scanners and sites, limiting the reproducibility of automated approaches. There is a need for multi-source annotated datasets to develop artificial intelligence-assisted fetal growth assessment methods. To address this bottleneck, we present an open, multi-centre, multi-device benchmark dataset of fetal US images with expert anatomical landmark annotations for clinically used fetal biometric measurements. These measurements include head bi-parietal and occipito-frontal diameters, abdominal transverse and antero-posterior diameters, and femoral length. The dataset comprises 4,513 de-identified US images from 1,904 subjects acquired at three clinical sites using seven different US devices. We provide standardised, subject-disjoint train/test splits, evaluation code, and baseline results to enable fair and reproducible comparison of methods. Using an automatic biometry model, we quantify domain shift and demonstrate that training and evaluation confined to a single centre substantially overestimate performance relative to multi-centre testing. To the best of our knowledge, this is the first publicly available multi-centre, multi-device, landmark-annotated dataset that covers all primary fetal biometry measures, providing a robust benchmark for domain adaptation and multi-centre generalisation in fetal biometry and enabling more reliable AI-assisted fetal growth assessment across centres. All data, annotations, training code, and evaluation pipelines are made publicly available.
SHADeS: Self-supervised Monocular Depth Estimation Through Non-Lambertian Image Decomposition
Feb 18, 2025Abstract:Purpose: Visual 3D scene reconstruction can support colonoscopy navigation. It can help in recognising which portions of the colon have been visualised and characterising the size and shape of polyps. This is still a very challenging problem due to complex illumination variations, including abundant specular reflections. We investigate how to effectively decouple light and depth in this problem. Methods: We introduce a self-supervised model that simultaneously characterises the shape and lighting of the visualised colonoscopy scene. Our model estimates shading, albedo, depth, and specularities (SHADeS) from single images. Unlike previous approaches (IID), we use a non-Lambertian model that treats specular reflections as a separate light component. The implementation of our method is available at https://github.com/RemaDaher/SHADeS. Results: We demonstrate on real colonoscopy images (Hyper Kvasir) that previous models for light decomposition (IID) and depth estimation (MonoVIT, ModoDepth2) are negatively affected by specularities. In contrast, SHADeS can simultaneously produce light decomposition and depth maps that are robust to specular regions. We also perform a quantitative comparison on phantom data (C3VD) where we further demonstrate the robustness of our model. Conclusion: Modelling specular reflections improves depth estimation in colonoscopy. We propose an effective self-supervised approach that uses this insight to jointly estimate light decomposition and depth. Light decomposition has the potential to help with other problems, such as place recognition within the colon.
SegCol Challenge: Semantic Segmentation for Tools and Fold Edges in Colonoscopy data
Dec 20, 2024

Abstract:Colorectal cancer (CRC) remains a leading cause of cancer-related deaths worldwide, with polyp removal being an effective early screening method. However, navigating the colon for thorough polyp detection poses significant challenges. To advance camera navigation in colonoscopy, we propose the Semantic Segmentation for Tools and Fold Edges in Colonoscopy (SegCol) Challenge. This challenge introduces a dataset from the EndoMapper repository, featuring manually annotated, pixel-level semantic labels for colon folds and endoscopic tools across selected frames from 96 colonoscopy videos. By providing fold edges as anatomical landmarks and depth discontinuity information from both fold and tool labels, the dataset is aimed to improve depth perception and localization methods. Hosted as part of the Endovis Challenge at MICCAI 2024, SegCol aims to drive innovation in colonoscopy navigation systems. Details are available at https://www.synapse.org/Synapse:syn54124209/wiki/626563, and code resources at https://github.com/surgical-vision/segcol_challenge .
SurgicalGS: Dynamic 3D Gaussian Splatting for Accurate Robotic-Assisted Surgical Scene Reconstruction
Oct 11, 2024



Abstract:Accurate 3D reconstruction of dynamic surgical scenes from endoscopic video is essential for robotic-assisted surgery. While recent 3D Gaussian Splatting methods have shown promise in achieving high-quality reconstructions with fast rendering speeds, their use of inverse depth loss functions compresses depth variations. This can lead to a loss of fine geometric details, limiting their ability to capture precise 3D geometry and effectiveness in intraoperative application. To address these challenges, we present SurgicalGS, a dynamic 3D Gaussian Splatting framework specifically designed for surgical scene reconstruction with improved geometric accuracy. Our approach first initialises a Gaussian point cloud using depth priors, employing binary motion masks to identify pixels with significant depth variations and fusing point clouds from depth maps across frames for initialisation. We use the Flexible Deformation Model to represent dynamic scene and introduce a normalised depth regularisation loss along with an unsupervised depth smoothness constraint to ensure more accurate geometric reconstruction. Extensive experiments on two real surgical datasets demonstrate that SurgicalGS achieves state-of-the-art reconstruction quality, especially in terms of accurate geometry, advancing the usability of 3D Gaussian Splatting in robotic-assisted surgery.
Tracking Everything in Robotic-Assisted Surgery
Sep 29, 2024Abstract:Accurate tracking of tissues and instruments in videos is crucial for Robotic-Assisted Minimally Invasive Surgery (RAMIS), as it enables the robot to comprehend the surgical scene with precise locations and interactions of tissues and tools. Traditional keypoint-based sparse tracking is limited by featured points, while flow-based dense two-view matching suffers from long-term drifts. Recently, the Tracking Any Point (TAP) algorithm was proposed to overcome these limitations and achieve dense accurate long-term tracking. However, its efficacy in surgical scenarios remains untested, largely due to the lack of a comprehensive surgical tracking dataset for evaluation. To address this gap, we introduce a new annotated surgical tracking dataset for benchmarking tracking methods for surgical scenarios, comprising real-world surgical videos with complex tissue and instrument motions. We extensively evaluate state-of-the-art (SOTA) TAP-based algorithms on this dataset and reveal their limitations in challenging surgical scenarios, including fast instrument motion, severe occlusions, and motion blur, etc. Furthermore, we propose a new tracking method, namely SurgMotion, to solve the challenges and further improve the tracking performance. Our proposed method outperforms most TAP-based algorithms in surgical instruments tracking, and especially demonstrates significant improvements over baselines in challenging medical videos.
PitVis-2023 Challenge: Workflow Recognition in videos of Endoscopic Pituitary Surgery
Sep 02, 2024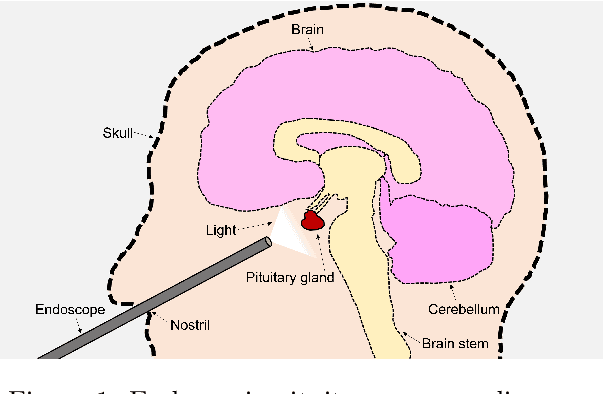
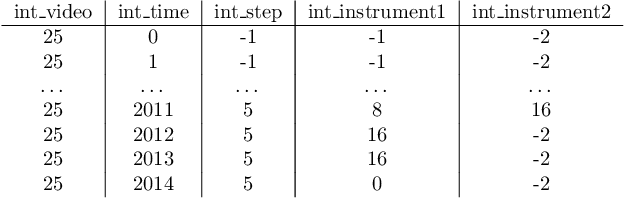
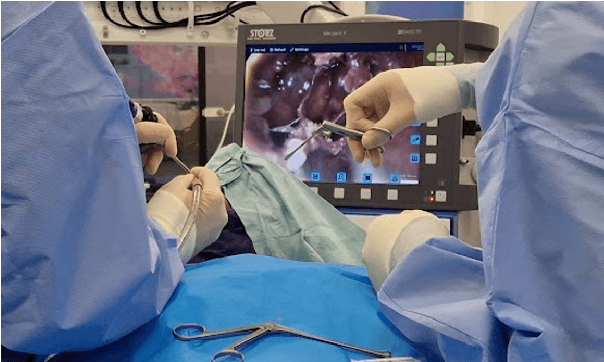
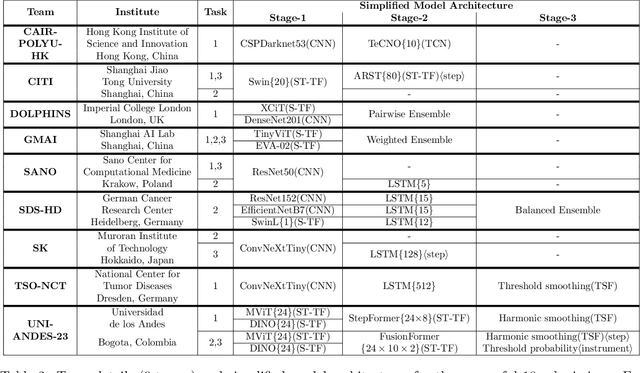
Abstract:The field of computer vision applied to videos of minimally invasive surgery is ever-growing. Workflow recognition pertains to the automated recognition of various aspects of a surgery: including which surgical steps are performed; and which surgical instruments are used. This information can later be used to assist clinicians when learning the surgery; during live surgery; and when writing operation notes. The Pituitary Vision (PitVis) 2023 Challenge tasks the community to step and instrument recognition in videos of endoscopic pituitary surgery. This is a unique task when compared to other minimally invasive surgeries due to the smaller working space, which limits and distorts vision; and higher frequency of instrument and step switching, which requires more precise model predictions. Participants were provided with 25-videos, with results presented at the MICCAI-2023 conference as part of the Endoscopic Vision 2023 Challenge in Vancouver, Canada, on 08-Oct-2023. There were 18-submissions from 9-teams across 6-countries, using a variety of deep learning models. A commonality between the top performing models was incorporating spatio-temporal and multi-task methods, with greater than 50% and 10% macro-F1-score improvement over purely spacial single-task models in step and instrument recognition respectively. The PitVis-2023 Challenge therefore demonstrates state-of-the-art computer vision models in minimally invasive surgery are transferable to a new dataset, with surgery specific techniques used to enhance performance, progressing the field further. Benchmark results are provided in the paper, and the dataset is publicly available at: https://doi.org/10.5522/04/26531686.
Mismatched: Evaluating the Limits of Image Matching Approaches and Benchmarks
Aug 29, 2024



Abstract:Three-dimensional (3D) reconstruction from two-dimensional images is an active research field in computer vision, with applications ranging from navigation and object tracking to segmentation and three-dimensional modeling. Traditionally, parametric techniques have been employed for this task. However, recent advancements have seen a shift towards learning-based methods. Given the rapid pace of research and the frequent introduction of new image matching methods, it is essential to evaluate them. In this paper, we present a comprehensive evaluation of various image matching methods using a structure-from-motion pipeline. We assess the performance of these methods on both in-domain and out-of-domain datasets, identifying key limitations in both the methods and benchmarks. We also investigate the impact of edge detection as a pre-processing step. Our analysis reveals that image matching for 3D reconstruction remains an open challenge, necessitating careful selection and tuning of models for specific scenarios, while also highlighting mismatches in how metrics currently represent method performance.
CathAction: A Benchmark for Endovascular Intervention Understanding
Aug 23, 2024Abstract:Real-time visual feedback from catheterization analysis is crucial for enhancing surgical safety and efficiency during endovascular interventions. However, existing datasets are often limited to specific tasks, small scale, and lack the comprehensive annotations necessary for broader endovascular intervention understanding. To tackle these limitations, we introduce CathAction, a large-scale dataset for catheterization understanding. Our CathAction dataset encompasses approximately 500,000 annotated frames for catheterization action understanding and collision detection, and 25,000 ground truth masks for catheter and guidewire segmentation. For each task, we benchmark recent related works in the field. We further discuss the challenges of endovascular intentions compared to traditional computer vision tasks and point out open research questions. We hope that CathAction will facilitate the development of endovascular intervention understanding methods that can be applied to real-world applications. The dataset is available at https://airvlab.github.io/cathdata/.
High-fidelity Endoscopic Image Synthesis by Utilizing Depth-guided Neural Surfaces
Apr 20, 2024Abstract:In surgical oncology, screening colonoscopy plays a pivotal role in providing diagnostic assistance, such as biopsy, and facilitating surgical navigation, particularly in polyp detection. Computer-assisted endoscopic surgery has recently gained attention and amalgamated various 3D computer vision techniques, including camera localization, depth estimation, surface reconstruction, etc. Neural Radiance Fields (NeRFs) and Neural Implicit Surfaces (NeuS) have emerged as promising methodologies for deriving accurate 3D surface models from sets of registered images, addressing the limitations of existing colon reconstruction approaches stemming from constrained camera movement. However, the inadequate tissue texture representation and confused scale problem in monocular colonoscopic image reconstruction still impede the progress of the final rendering results. In this paper, we introduce a novel method for colon section reconstruction by leveraging NeuS applied to endoscopic images, supplemented by a single frame of depth map. Notably, we pioneered the exploration of utilizing only one frame depth map in photorealistic reconstruction and neural rendering applications while this single depth map can be easily obtainable from other monocular depth estimation networks with an object scale. Through rigorous experimentation and validation on phantom imagery, our approach demonstrates exceptional accuracy in completely rendering colon sections, even capturing unseen portions of the surface. This breakthrough opens avenues for achieving stable and consistently scaled reconstructions, promising enhanced quality in cancer screening procedures and treatment interventions.
Measuring proximity to standard planes during fetal brain ultrasound scanning
Apr 10, 2024



Abstract:This paper introduces a novel pipeline designed to bring ultrasound (US) plane pose estimation closer to clinical use for more effective navigation to the standard planes (SPs) in the fetal brain. We propose a semi-supervised segmentation model utilizing both labeled SPs and unlabeled 3D US volume slices. Our model enables reliable segmentation across a diverse set of fetal brain images. Furthermore, the model incorporates a classification mechanism to identify the fetal brain precisely. Our model not only filters out frames lacking the brain but also generates masks for those containing it, enhancing the relevance of plane pose regression in clinical settings. We focus on fetal brain navigation from 2D ultrasound (US) video analysis and combine this model with a US plane pose regression network to provide sensorless proximity detection to SPs and non-SPs planes; we emphasize the importance of proximity detection to SPs for guiding sonographers, offering a substantial advantage over traditional methods by allowing earlier and more precise adjustments during scanning. We demonstrate the practical applicability of our approach through validation on real fetal scan videos obtained from sonographers of varying expertise levels. Our findings demonstrate the potential of our approach to complement existing fetal US technologies and advance prenatal diagnostic practices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge