Jialei Chen
I2V-GS: Infrastructure-to-Vehicle View Transformation with Gaussian Splatting for Autonomous Driving Data Generation
Jul 31, 2025Abstract:Vast and high-quality data are essential for end-to-end autonomous driving systems. However, current driving data is mainly collected by vehicles, which is expensive and inefficient. A potential solution lies in synthesizing data from real-world images. Recent advancements in 3D reconstruction demonstrate photorealistic novel view synthesis, highlighting the potential of generating driving data from images captured on the road. This paper introduces a novel method, I2V-GS, to transfer the Infrastructure view To the Vehicle view with Gaussian Splatting. Reconstruction from sparse infrastructure viewpoints and rendering under large view transformations is a challenging problem. We adopt the adaptive depth warp to generate dense training views. To further expand the range of views, we employ a cascade strategy to inpaint warped images, which also ensures inpainting content is consistent across views. To further ensure the reliability of the diffusion model, we utilize the cross-view information to perform a confidenceguided optimization. Moreover, we introduce RoadSight, a multi-modality, multi-view dataset from real scenarios in infrastructure views. To our knowledge, I2V-GS is the first framework to generate autonomous driving datasets with infrastructure-vehicle view transformation. Experimental results demonstrate that I2V-GS significantly improves synthesis quality under vehicle view, outperforming StreetGaussian in NTA-Iou, NTL-Iou, and FID by 45.7%, 34.2%, and 14.9%, respectively.
Unlocking the Power of Diffusion Models in Sequential Recommendation: A Simple and Effective Approach
May 26, 2025



Abstract:In this paper, we focus on the often-overlooked issue of embedding collapse in existing diffusion-based sequential recommendation models and propose ADRec, an innovative framework designed to mitigate this problem. Diverging from previous diffusion-based methods, ADRec applies an independent noise process to each token and performs diffusion across the entire target sequence during training. ADRec captures token interdependency through auto-regression while modeling per-token distributions through token-level diffusion. This dual approach enables the model to effectively capture both sequence dynamics and item representations, overcoming the limitations of existing methods. To further mitigate embedding collapse, we propose a three-stage training strategy: (1) pre-training the embedding weights, (2) aligning these weights with the ADRec backbone, and (3) fine-tuning the model. During inference, ADRec applies the denoising process only to the last token, ensuring that the meaningful patterns in historical interactions are preserved. Our comprehensive empirical evaluation across six datasets underscores the effectiveness of ADRec in enhancing both the accuracy and efficiency of diffusion-based sequential recommendation systems.
MLLMs are Deeply Affected by Modality Bias
May 24, 2025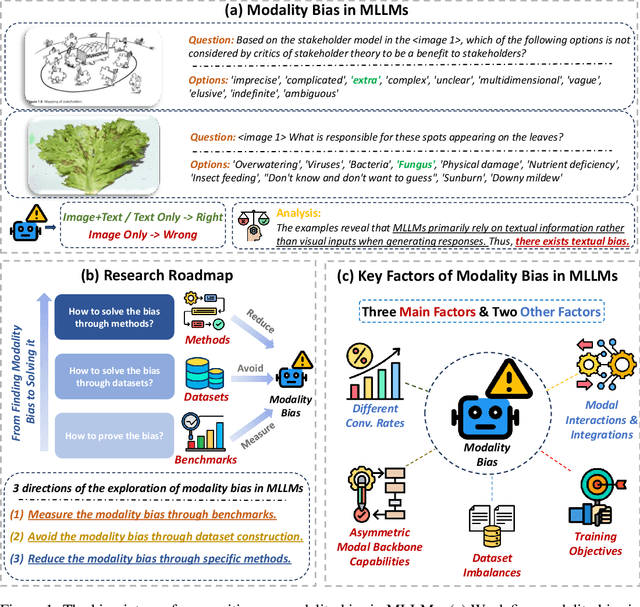
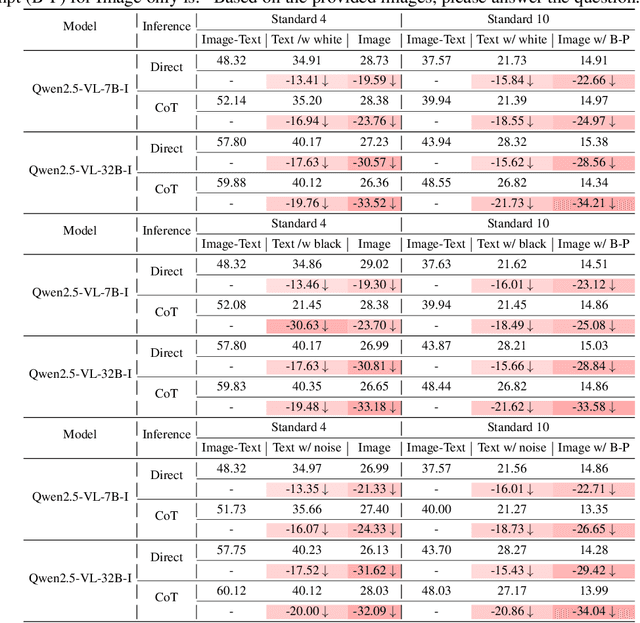
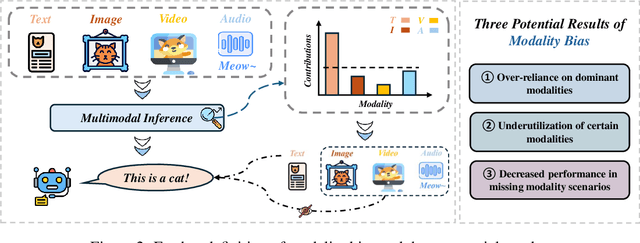
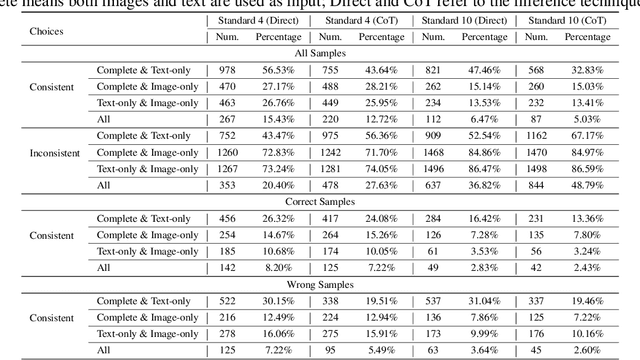
Abstract:Recent advances in Multimodal Large Language Models (MLLMs) have shown promising results in integrating diverse modalities such as texts and images. MLLMs are heavily influenced by modality bias, often relying on language while under-utilizing other modalities like visual inputs. This position paper argues that MLLMs are deeply affected by modality bias. Firstly, we diagnose the current state of modality bias, highlighting its manifestations across various tasks. Secondly, we propose a systematic research road-map related to modality bias in MLLMs. Thirdly, we identify key factors of modality bias in MLLMs and offer actionable suggestions for future research to mitigate it. To substantiate these findings, we conduct experiments that demonstrate the influence of each factor: 1. Data Characteristics: Language data is compact and abstract, while visual data is redundant and complex, creating an inherent imbalance in learning dynamics. 2. Imbalanced Backbone Capabilities: The dominance of pretrained language models in MLLMs leads to overreliance on language and neglect of visual information. 3. Training Objectives: Current objectives often fail to promote balanced cross-modal alignment, resulting in shortcut learning biased toward language. These findings highlight the need for balanced training strategies and model architectures to better integrate multiple modalities in MLLMs. We call for interdisciplinary efforts to tackle these challenges and drive innovation in MLLM research. Our work provides a fresh perspective on modality bias in MLLMs and offers insights for developing more robust and generalizable multimodal systems-advancing progress toward Artificial General Intelligence.
Split Matching for Inductive Zero-shot Semantic Segmentation
May 08, 2025Abstract:Zero-shot Semantic Segmentation (ZSS) aims to segment categories that are not annotated during training. While fine-tuning vision-language models has achieved promising results, these models often overfit to seen categories due to the lack of supervision for unseen classes. As an alternative to fully supervised approaches, query-based segmentation has shown great latent in ZSS, as it enables object localization without relying on explicit labels. However, conventional Hungarian matching, a core component in query-based frameworks, needs full supervision and often misclassifies unseen categories as background in the setting of ZSS. To address this issue, we propose Split Matching (SM), a novel assignment strategy that decouples Hungarian matching into two components: one for seen classes in annotated regions and another for latent classes in unannotated regions (referred to as unseen candidates). Specifically, we partition the queries into seen and candidate groups, enabling each to be optimized independently according to its available supervision. To discover unseen candidates, we cluster CLIP dense features to generate pseudo masks and extract region-level embeddings using CLS tokens. Matching is then conducted separately for the two groups based on both class-level similarity and mask-level consistency. Additionally, we introduce a Multi-scale Feature Enhancement (MFE) module that refines decoder features through residual multi-scale aggregation, improving the model's ability to capture spatial details across resolutions. SM is the first to introduce decoupled Hungarian matching under the inductive ZSS setting, and achieves state-of-the-art performance on two standard benchmarks.
OmniSAM: Omnidirectional Segment Anything Model for UDA in Panoramic Semantic Segmentation
Mar 10, 2025Abstract:Segment Anything Model 2 (SAM2) has emerged as a strong base model in various pinhole imaging segmentation tasks. However, when applying it to $360^\circ$ domain, the significant field-of-view (FoV) gap between pinhole ($70^\circ \times 70^\circ$) and panoramic images ($180^\circ \times 360^\circ$) poses unique challenges. Two major concerns for this application includes 1) inevitable distortion and object deformation brought by the large FoV disparity between domains; 2) the lack of pixel-level semantic understanding that the original SAM2 cannot provide. To address these issues, we propose a novel OmniSAM framework, which makes the first attempt to apply SAM2 for panoramic semantic segmentation. Specifically, to bridge the first gap, OmniSAM first divides the panorama into sequences of patches. These patches are then treated as image sequences in similar manners as in video segmentation tasks. We then leverage the SAM2's memory mechanism to extract cross-patch correspondences that embeds the cross-FoV dependencies, improving feature continuity and the prediction consistency along mask boundaries. For the second gap, OmniSAM fine-tunes the pretrained image encoder and reutilize the mask decoder for semantic prediction. An FoV-based prototypical adaptation module with dynamic pseudo label update mechanism is also introduced to facilitate the alignment of memory and backbone features, thereby improving model generalization ability across different sizes of source models. Extensive experimental results demonstrate that OmniSAM outperforms the state-of-the-art methods by large margins, e.g., 79.06% (+10.22%) on SPin8-to-SPan8, 62.46% (+6.58%) on CS13-to-DP13.
Learning Robust Anymodal Segmentor with Unimodal and Cross-modal Distillation
Nov 26, 2024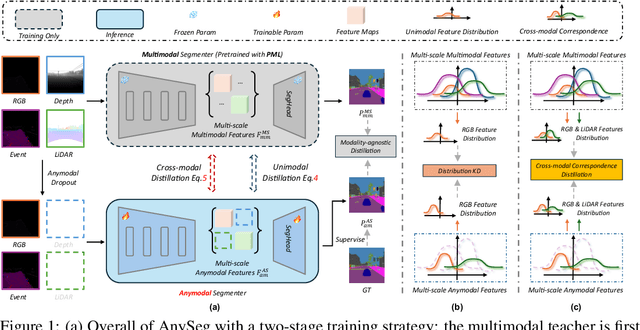
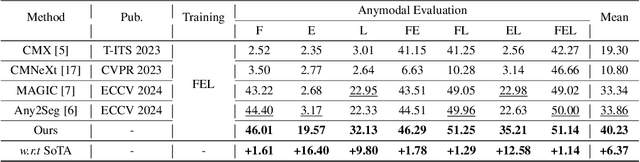
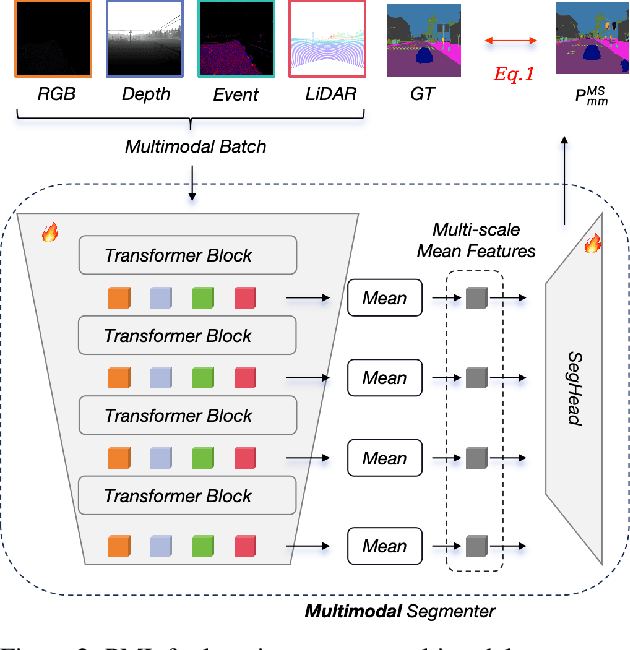
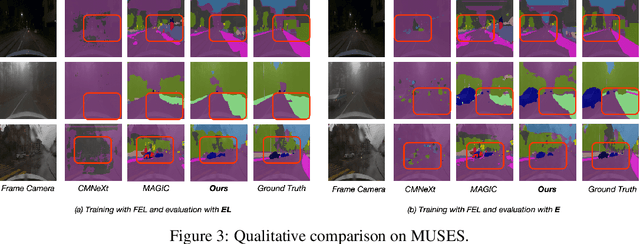
Abstract:Simultaneously using multimodal inputs from multiple sensors to train segmentors is intuitively advantageous but practically challenging. A key challenge is unimodal bias, where multimodal segmentors over rely on certain modalities, causing performance drops when others are missing, common in real world applications. To this end, we develop the first framework for learning robust segmentor that can handle any combinations of visual modalities. Specifically, we first introduce a parallel multimodal learning strategy for learning a strong teacher. The cross-modal and unimodal distillation is then achieved in the multi scale representation space by transferring the feature level knowledge from multimodal to anymodal segmentors, aiming at addressing the unimodal bias and avoiding over-reliance on specific modalities. Moreover, a prediction level modality agnostic semantic distillation is proposed to achieve semantic knowledge transferring for segmentation. Extensive experiments on both synthetic and real-world multi-sensor benchmarks demonstrate that our method achieves superior performance.
SurgicalGS: Dynamic 3D Gaussian Splatting for Accurate Robotic-Assisted Surgical Scene Reconstruction
Oct 11, 2024



Abstract:Accurate 3D reconstruction of dynamic surgical scenes from endoscopic video is essential for robotic-assisted surgery. While recent 3D Gaussian Splatting methods have shown promise in achieving high-quality reconstructions with fast rendering speeds, their use of inverse depth loss functions compresses depth variations. This can lead to a loss of fine geometric details, limiting their ability to capture precise 3D geometry and effectiveness in intraoperative application. To address these challenges, we present SurgicalGS, a dynamic 3D Gaussian Splatting framework specifically designed for surgical scene reconstruction with improved geometric accuracy. Our approach first initialises a Gaussian point cloud using depth priors, employing binary motion masks to identify pixels with significant depth variations and fusing point clouds from depth maps across frames for initialisation. We use the Flexible Deformation Model to represent dynamic scene and introduce a normalised depth regularisation loss along with an unsupervised depth smoothness constraint to ensure more accurate geometric reconstruction. Extensive experiments on two real surgical datasets demonstrate that SurgicalGS achieves state-of-the-art reconstruction quality, especially in terms of accurate geometry, advancing the usability of 3D Gaussian Splatting in robotic-assisted surgery.
Generalizable Semantic Vision Query Generation for Zero-shot Panoptic and Semantic Segmentation
Feb 21, 2024



Abstract:Zero-shot Panoptic Segmentation (ZPS) aims to recognize foreground instances and background stuff without images containing unseen categories in training. Due to the visual data sparsity and the difficulty of generalizing from seen to unseen categories, this task remains challenging. To better generalize to unseen classes, we propose Conditional tOken aligNment and Cycle trAnsiTion (CONCAT), to produce generalizable semantic vision queries. First, a feature extractor is trained by CON to link the vision and semantics for providing target queries. Formally, CON is proposed to align the semantic queries with the CLIP visual CLS token extracted from complete and masked images. To address the lack of unseen categories, a generator is required. However, one of the gaps in synthesizing pseudo vision queries, ie, vision queries for unseen categories, is describing fine-grained visual details through semantic embeddings. Therefore, we approach CAT to train the generator in semantic-vision and vision-semantic manners. In semantic-vision, visual query contrast is proposed to model the high granularity of vision by pulling the pseudo vision queries with the corresponding targets containing segments while pushing those without segments away. To ensure the generated queries retain semantic information, in vision-semantic, the pseudo vision queries are mapped back to semantic and supervised by real semantic embeddings. Experiments on ZPS achieve a 5.2% hPQ increase surpassing SOTA. We also examine inductive ZPS and open-vocabulary semantic segmentation and obtain comparative results while being 2 times faster in testing.
CLIP Is Also a Good Teacher: A New Learning Framework for Inductive Zero-shot Semantic Segmentation
Oct 03, 2023Abstract:Existing Generalized Zero-shot Semantic Segmentation (GZLSS) methods apply either finetuning the CLIP paradigm or formulating it as a mask classification task, benefiting from the Vision-Language Models (VLMs). However, the fine-tuning methods are restricted with fixed backbone models which are not flexible for segmentation, and mask classification methods heavily rely on additional explicit mask proposers. Meanwhile, prevalent methods utilize only seen categories which is a great waste, i.e., neglecting the area exists but not annotated. To this end, we propose CLIPTeacher, a new learning framework that can be applied to various per-pixel classification segmentation models without introducing any explicit mask proposer or changing the structure of CLIP, and utilize both seen and ignoring areas. Specifically, CLIPTeacher consists of two key modules: Global Learning Module (GLM) and Pixel Learning Module (PLM). Specifically, GLM aligns the dense features from an image encoder with the CLS token, i.e., the only token trained in CLIP, which is a simple but effective way to probe global information from the CLIP models. In contrast, PLM only leverages dense tokens from CLIP to produce high-level pseudo annotations for ignoring areas without introducing any extra mask proposer. Meanwhile, PLM can fully take advantage of the whole image based on the pseudo annotations. Experimental results on three benchmark datasets: PASCAL VOC 2012, COCO-Stuff 164k, and PASCAL Context show large performance gains, i.e., 2.2%, 1.3%, and 8.8%
Deep Generative Imputation Model for Missing Not At Random Data
Aug 16, 2023



Abstract:Data analysis usually suffers from the Missing Not At Random (MNAR) problem, where the cause of the value missing is not fully observed. Compared to the naive Missing Completely At Random (MCAR) problem, it is more in line with the realistic scenario whereas more complex and challenging. Existing statistical methods model the MNAR mechanism by different decomposition of the joint distribution of the complete data and the missing mask. But we empirically find that directly incorporating these statistical methods into deep generative models is sub-optimal. Specifically, it would neglect the confidence of the reconstructed mask during the MNAR imputation process, which leads to insufficient information extraction and less-guaranteed imputation quality. In this paper, we revisit the MNAR problem from a novel perspective that the complete data and missing mask are two modalities of incomplete data on an equal footing. Along with this line, we put forward a generative-model-specific joint probability decomposition method, conjunction model, to represent the distributions of two modalities in parallel and extract sufficient information from both complete data and missing mask. Taking a step further, we exploit a deep generative imputation model, namely GNR, to process the real-world missing mechanism in the latent space and concurrently impute the incomplete data and reconstruct the missing mask. The experimental results show that our GNR surpasses state-of-the-art MNAR baselines with significant margins (averagely improved from 9.9% to 18.8% in RMSE) and always gives a better mask reconstruction accuracy which makes the imputation more principle.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge