Daniel S. Elson
SAMSA: Segment Anything Model Enhanced with Spectral Angles for Hyperspectral Interactive Medical Image Segmentation
Jul 31, 2025Abstract:Hyperspectral imaging (HSI) provides rich spectral information for medical imaging, yet encounters significant challenges due to data limitations and hardware variations. We introduce SAMSA, a novel interactive segmentation framework that combines an RGB foundation model with spectral analysis. SAMSA efficiently utilizes user clicks to guide both RGB segmentation and spectral similarity computations. The method addresses key limitations in HSI segmentation through a unique spectral feature fusion strategy that operates independently of spectral band count and resolution. Performance evaluation on publicly available datasets has shown 81.0% 1-click and 93.4% 5-click DICE on a neurosurgical and 81.1% 1-click and 89.2% 5-click DICE on an intraoperative porcine hyperspectral dataset. Experimental results demonstrate SAMSA's effectiveness in few-shot and zero-shot learning scenarios and using minimal training examples. Our approach enables seamless integration of datasets with different spectral characteristics, providing a flexible framework for hyperspectral medical image analysis.
Hybrid Deep Reinforcement Learning for Radio Tracer Localisation in Robotic-assisted Radioguided Surgery
Mar 11, 2025



Abstract:Radioguided surgery, such as sentinel lymph node biopsy, relies on the precise localization of radioactive targets by non-imaging gamma/beta detectors. Manual radioactive target detection based on visual display or audible indication of gamma level is highly dependent on the ability of the surgeon to track and interpret the spatial information. This paper presents a learning-based method to realize the autonomous radiotracer detection in robot-assisted surgeries by navigating the probe to the radioactive target. We proposed novel hybrid approach that combines deep reinforcement learning (DRL) with adaptive robotic scanning. The adaptive grid-based scanning could provide initial direction estimation while the DRL-based agent could efficiently navigate to the target utilising historical data. Simulation experiments demonstrate a 95% success rate, and improved efficiency and robustness compared to conventional techniques. Real-world evaluation on the da Vinci Research Kit (dVRK) further confirms the feasibility of the approach, achieving an 80% success rate in radiotracer detection. This method has the potential to enhance consistency, reduce operator dependency, and improve procedural accuracy in radioguided surgeries.
Automatic Robotic-Assisted Diffuse Reflectance Spectroscopy Scanning System
Mar 11, 2025



Abstract:Diffuse Reflectance Spectroscopy (DRS) is a well-established optical technique for tissue composition assessment which has been clinically evaluated for tumour detection to ensure the complete removal of cancerous tissue. While point-wise assessment has many potential applications, incorporating automated large-area scanning would enable holistic tissue sampling with higher consistency. We propose a robotic system to facilitate autonomous DRS scanning with hybrid visual servoing control. A specially designed height compensation module enables precise contact condition control. The evaluation results show that the system can accurately execute the scanning command and acquire consistent DRS spectra with comparable results to the manual collection, which is the current gold standard protocol. Integrating the proposed system into surgery lays the groundwork for autonomous intra-operative DRS tissue assessment with high reliability and repeatability. This could reduce the need for manual scanning by the surgeon while ensuring complete tumor removal in clinical practice.
Memorized action chunking with Transformers: Imitation learning for vision-based tissue surface scanning
Nov 06, 2024
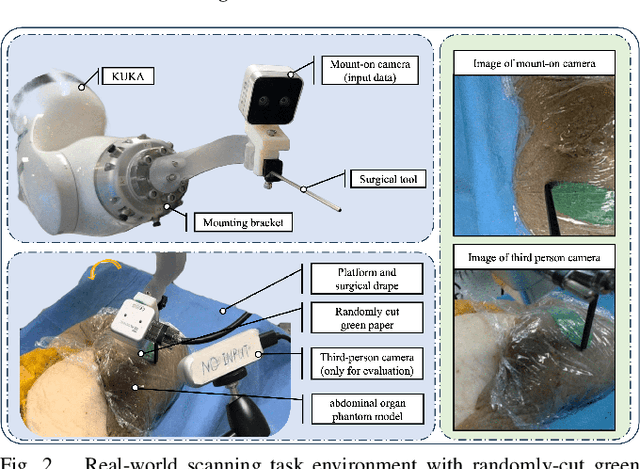
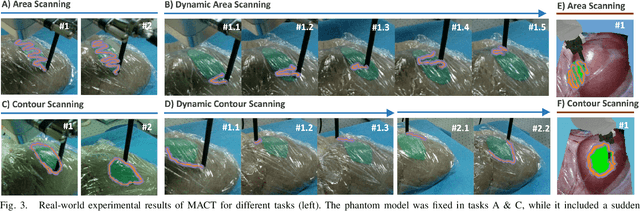

Abstract:Optical sensing technologies are emerging technologies used in cancer surgeries to ensure the complete removal of cancerous tissue. While point-wise assessment has many potential applications, incorporating automated large area scanning would enable holistic tissue sampling. However, such scanning tasks are challenging due to their long-horizon dependency and the requirement for fine-grained motion. To address these issues, we introduce Memorized Action Chunking with Transformers (MACT), an intuitive yet efficient imitation learning method for tissue surface scanning tasks. It utilizes a sequence of past images as historical information to predict near-future action sequences. In addition, hybrid temporal-spatial positional embeddings were employed to facilitate learning. In various simulation settings, MACT demonstrated significant improvements in contour scanning and area scanning over the baseline model. In real-world testing, with only 50 demonstration trajectories, MACT surpassed the baseline model by achieving a 60-80% success rate on all scanning tasks. Our findings suggest that MACT is a promising model for adaptive scanning in surgical settings.
SurgicalGS: Dynamic 3D Gaussian Splatting for Accurate Robotic-Assisted Surgical Scene Reconstruction
Oct 11, 2024



Abstract:Accurate 3D reconstruction of dynamic surgical scenes from endoscopic video is essential for robotic-assisted surgery. While recent 3D Gaussian Splatting methods have shown promise in achieving high-quality reconstructions with fast rendering speeds, their use of inverse depth loss functions compresses depth variations. This can lead to a loss of fine geometric details, limiting their ability to capture precise 3D geometry and effectiveness in intraoperative application. To address these challenges, we present SurgicalGS, a dynamic 3D Gaussian Splatting framework specifically designed for surgical scene reconstruction with improved geometric accuracy. Our approach first initialises a Gaussian point cloud using depth priors, employing binary motion masks to identify pixels with significant depth variations and fusing point clouds from depth maps across frames for initialisation. We use the Flexible Deformation Model to represent dynamic scene and introduce a normalised depth regularisation loss along with an unsupervised depth smoothness constraint to ensure more accurate geometric reconstruction. Extensive experiments on two real surgical datasets demonstrate that SurgicalGS achieves state-of-the-art reconstruction quality, especially in terms of accurate geometry, advancing the usability of 3D Gaussian Splatting in robotic-assisted surgery.
Tracking Everything in Robotic-Assisted Surgery
Sep 29, 2024Abstract:Accurate tracking of tissues and instruments in videos is crucial for Robotic-Assisted Minimally Invasive Surgery (RAMIS), as it enables the robot to comprehend the surgical scene with precise locations and interactions of tissues and tools. Traditional keypoint-based sparse tracking is limited by featured points, while flow-based dense two-view matching suffers from long-term drifts. Recently, the Tracking Any Point (TAP) algorithm was proposed to overcome these limitations and achieve dense accurate long-term tracking. However, its efficacy in surgical scenarios remains untested, largely due to the lack of a comprehensive surgical tracking dataset for evaluation. To address this gap, we introduce a new annotated surgical tracking dataset for benchmarking tracking methods for surgical scenarios, comprising real-world surgical videos with complex tissue and instrument motions. We extensively evaluate state-of-the-art (SOTA) TAP-based algorithms on this dataset and reveal their limitations in challenging surgical scenarios, including fast instrument motion, severe occlusions, and motion blur, etc. Furthermore, we propose a new tracking method, namely SurgMotion, to solve the challenges and further improve the tracking performance. Our proposed method outperforms most TAP-based algorithms in surgical instruments tracking, and especially demonstrates significant improvements over baselines in challenging medical videos.
Detecting the Sensing Area of A Laparoscopic Probe in Minimally Invasive Cancer Surgery
Jul 07, 2023



Abstract:In surgical oncology, it is challenging for surgeons to identify lymph nodes and completely resect cancer even with pre-operative imaging systems like PET and CT, because of the lack of reliable intraoperative visualization tools. Endoscopic radio-guided cancer detection and resection has recently been evaluated whereby a novel tethered laparoscopic gamma detector is used to localize a preoperatively injected radiotracer. This can both enhance the endoscopic imaging and complement preoperative nuclear imaging data. However, gamma activity visualization is challenging to present to the operator because the probe is non-imaging and it does not visibly indicate the activity origination on the tissue surface. Initial failed attempts used segmentation or geometric methods, but led to the discovery that it could be resolved by leveraging high-dimensional image features and probe position information. To demonstrate the effectiveness of this solution, we designed and implemented a simple regression network that successfully addressed the problem. To further validate the proposed solution, we acquired and publicly released two datasets captured using a custom-designed, portable stereo laparoscope system. Through intensive experimentation, we demonstrated that our method can successfully and effectively detect the sensing area, establishing a new performance benchmark. Code and data are available at https://github.com/br0202/Sensing_area_detection.git
Deep Imitation Learning for Automated Drop-In Gamma Probe Manipulation
Apr 27, 2023Abstract:The increasing prevalence of prostate cancer has led to the widespread adoption of Robotic-Assisted Surgery (RAS) as a treatment option. Sentinel lymph node biopsy (SLNB) is a crucial component of prostate cancer surgery and requires accurate diagnostic evidence. This procedure can be improved by using a drop-in gamma probe, SENSEI system, to distinguish cancerous tissue from normal tissue. However, manual control of the probe using live gamma level display and audible feedback could be challenging for inexperienced surgeons, leading to the potential for missed detections. In this study, a deep imitation training workflow was proposed to automate the radioactive node detection procedure. The proposed training workflow uses simulation data to train an end-to-end vision-based gamma probe manipulation agent. The evaluation results showed that the proposed approach was capable to predict the next-step action and holds promise for further improvement and extension to a hardware setup.
Self-Supervised Depth Estimation in Laparoscopic Image using 3D Geometric Consistency
Aug 17, 2022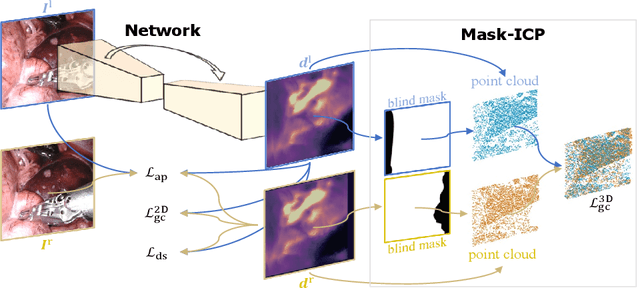
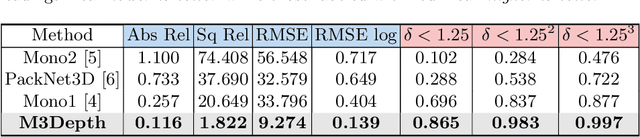
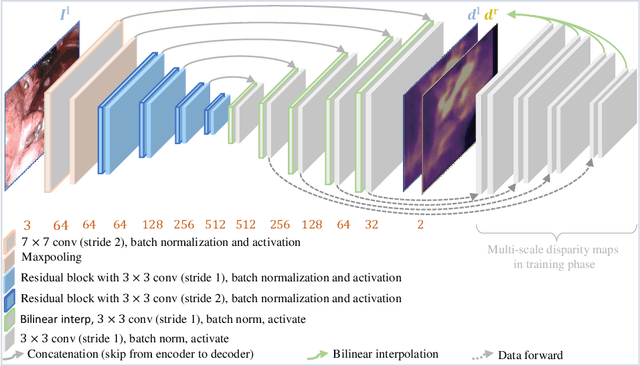
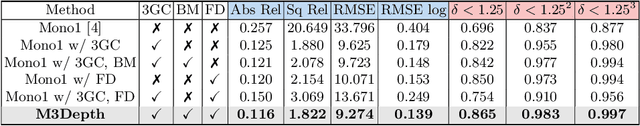
Abstract:Depth estimation is a crucial step for image-guided intervention in robotic surgery and laparoscopic imaging system. Since per-pixel depth ground truth is difficult to acquire for laparoscopic image data, it is rarely possible to apply supervised depth estimation to surgical applications. As an alternative, self-supervised methods have been introduced to train depth estimators using only synchronized stereo image pairs. However, most recent work focused on the left-right consistency in 2D and ignored valuable inherent 3D information on the object in real world coordinates, meaning that the left-right 3D geometric structural consistency is not fully utilized. To overcome this limitation, we present M3Depth, a self-supervised depth estimator to leverage 3D geometric structural information hidden in stereo pairs while keeping monocular inference. The method also removes the influence of border regions unseen in at least one of the stereo images via masking, to enhance the correspondences between left and right images in overlapping areas. Intensive experiments show that our method outperforms previous self-supervised approaches on both a public dataset and a newly acquired dataset by a large margin, indicating a good generalization across different samples and laparoscopes.
Self-Supervised Generative Adversarial Network for Depth Estimation in Laparoscopic Images
Jul 09, 2021
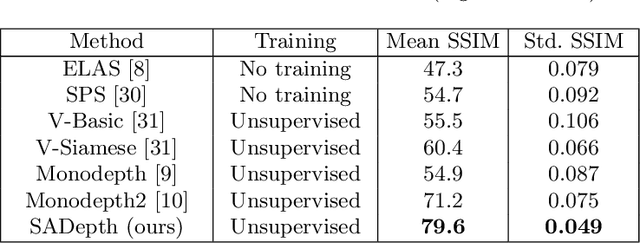
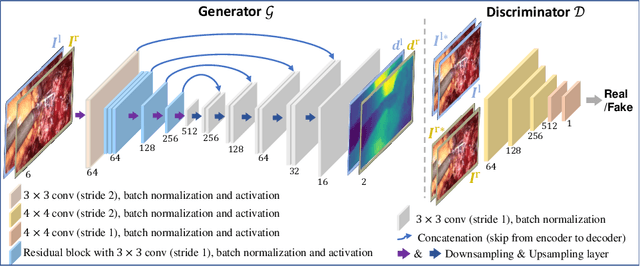
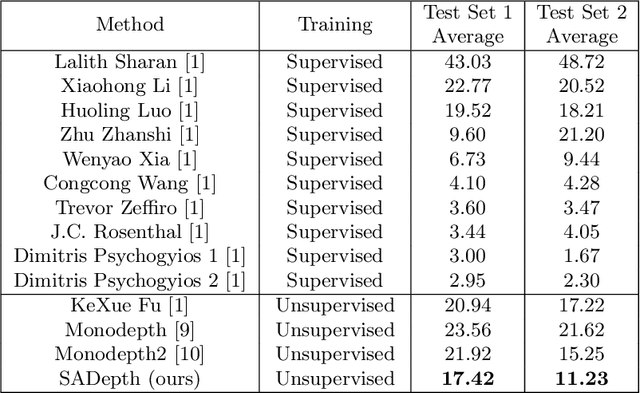
Abstract:Dense depth estimation and 3D reconstruction of a surgical scene are crucial steps in computer assisted surgery. Recent work has shown that depth estimation from a stereo images pair could be solved with convolutional neural networks. However, most recent depth estimation models were trained on datasets with per-pixel ground truth. Such data is especially rare for laparoscopic imaging, making it hard to apply supervised depth estimation to real surgical applications. To overcome this limitation, we propose SADepth, a new self-supervised depth estimation method based on Generative Adversarial Networks. It consists of an encoder-decoder generator and a discriminator to incorporate geometry constraints during training. Multi-scale outputs from the generator help to solve the local minima caused by the photometric reprojection loss, while the adversarial learning improves the framework generation quality. Extensive experiments on two public datasets show that SADepth outperforms recent state-of-the-art unsupervised methods by a large margin, and reduces the gap between supervised and unsupervised depth estimation in laparoscopic images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge