Jian-Qing Zheng
Optical Computation-in-Communication enables low-latency, high-fidelity perception in telesurgery
Oct 15, 2025Abstract:Artificial intelligence (AI) holds significant promise for enhancing intraoperative perception and decision-making in telesurgery, where physical separation impairs sensory feedback and control. Despite advances in medical AI and surgical robotics, conventional electronic AI architectures remain fundamentally constrained by the compounded latency from serial processing of inference and communication. This limitation is especially critical in latency-sensitive procedures such as endovascular interventions, where delays over 200 ms can compromise real-time AI reliability and patient safety. Here, we introduce an Optical Computation-in-Communication (OCiC) framework that reduces end-to-end latency significantly by performing AI inference concurrently with optical communication. OCiC integrates Optical Remote Computing Units (ORCUs) directly into the optical communication pathway, with each ORCU experimentally achieving up to 69 tera-operations per second per channel through spectrally efficient two-dimensional photonic convolution. The system maintains ultrahigh inference fidelity within 0.1% of CPU/GPU baselines on classification and coronary angiography segmentation, while intrinsically mitigating cumulative error propagation, a longstanding barrier to deep optical network scalability. We validated the robustness of OCiC through outdoor dark fibre deployments, confirming consistent and stable performance across varying environmental conditions. When scaled globally, OCiC transforms long-haul fibre infrastructure into a distributed photonic AI fabric with exascale potential, enabling reliable, low-latency telesurgery across distances up to 10,000 km and opening a new optical frontier for distributed medical intelligence.
Deformation-Recovery Diffusion Model (DRDM): Instance Deformation for Image Manipulation and Synthesis
Jul 10, 2024Abstract:In medical imaging, the diffusion models have shown great potential in synthetic image generation tasks. However, these models often struggle with the interpretable connections between the generated and existing images and could create illusions. To address these challenges, our research proposes a novel diffusion-based generative model based on deformation diffusion and recovery. This model, named Deformation-Recovery Diffusion Model (DRDM), diverges from traditional score/intensity and latent feature-based approaches, emphasizing morphological changes through deformation fields rather than direct image synthesis. This is achieved by introducing a topological-preserving deformation field generation method, which randomly samples and integrates a set of multi-scale Deformation Vector Fields (DVF). DRDM is trained to learn to recover unreasonable deformation components, thereby restoring each randomly deformed image to a realistic distribution. These innovations facilitate the generation of diverse and anatomically plausible deformations, enhancing data augmentation and synthesis for further analysis in downstream tasks, such as few-shot learning and image registration. Experimental results in cardiac MRI and pulmonary CT show DRDM is capable of creating diverse, large (over 10% image size deformation scale), and high-quality (negative ratio of folding rate is lower than 1%) deformation fields. The further experimental results in downstream tasks, 2D image segmentation and 3D image registration, indicate significant improvements resulting from DRDM, showcasing the potential of our model to advance image manipulation and synthesis in medical imaging and beyond. Our implementation will be available at https://github.com/jianqingzheng/def_diff_rec.
VMambaMorph: a Multi-Modality Deformable Image Registration Framework based on Visual State Space Model with Cross-Scan Module
Apr 14, 2024


Abstract:Image registration, a critical process in medical imaging, involves aligning different sets of medical imaging data into a single unified coordinate system. Deep learning networks, such as the Convolutional Neural Network (CNN)-based VoxelMorph, Vision Transformer (ViT)-based TransMorph, and State Space Model (SSM)-based MambaMorph, have demonstrated effective performance in this domain. The recent Visual State Space Model (VMamba), which incorporates a cross-scan module with SSM, has exhibited promising improvements in modeling global-range dependencies with efficient computational cost in computer vision tasks. This paper hereby introduces an exploration of VMamba with image registration, named VMambaMorph. This novel hybrid VMamba-CNN network is designed specifically for 3D image registration. Utilizing a U-shaped network architecture, VMambaMorph computes the deformation field based on target and source volumes. The VMamba-based block with 2D cross-scan module is redesigned for 3D volumetric feature processing. To overcome the complex motion and structure on multi-modality images, we further propose a fine-tune recursive registration framework. We validate VMambaMorph using a public benchmark brain MR-CT registration dataset, comparing its performance against current state-of-the-art methods. The results indicate that VMambaMorph achieves competitive registration quality. The code for VMambaMorph with all baseline methods is available on GitHub.
Mamba-UNet: UNet-Like Pure Visual Mamba for Medical Image Segmentation
Feb 07, 2024Abstract:In recent advancements in medical image analysis, Convolutional Neural Networks (CNN) and Vision Transformers (ViT) have set significant benchmarks. While the former excels in capturing local features through its convolution operations, the latter achieves remarkable global context understanding by leveraging self-attention mechanisms. However, both architectures exhibit limitations in efficiently modeling long-range dependencies within medical images, which is a critical aspect for precise segmentation. Inspired by the Mamba architecture, known for its proficiency in handling long sequences and global contextual information with enhanced computational efficiency as a State Space Model (SSM), we propose Mamba-UNet, a novel architecture that synergizes the U-Net in medical image segmentation with Mamba's capability. Mamba-UNet adopts a pure Visual Mamba (VMamba)-based encoder-decoder structure, infused with skip connections to preserve spatial information across different scales of the network. This design facilitates a comprehensive feature learning process, capturing intricate details and broader semantic contexts within medical images. We introduce a novel integration mechanism within the VMamba blocks to ensure seamless connectivity and information flow between the encoder and decoder paths, enhancing the segmentation performance. We conducted experiments on publicly available MRI cardiac multi-structures segmentation dataset. The results show that Mamba-UNet outperforms UNet, Swin-UNet in medical image segmentation under the same hyper-parameter setting. The source code and baseline implementations are available.
MGDepth: Motion-Guided Cost Volume For Self-Supervised Monocular Depth In Dynamic Scenarios
Dec 23, 2023



Abstract:Despite advancements in self-supervised monocular depth estimation, challenges persist in dynamic scenarios due to the dependence on assumptions about a static world. In this paper, we present MGDepth, a Motion-Guided Cost Volume Depth Net, to achieve precise depth estimation for both dynamic objects and static backgrounds, all while maintaining computational efficiency. To tackle the challenges posed by dynamic content, we incorporate optical flow and coarse monocular depth to create a novel static reference frame. This frame is then utilized to build a motion-guided cost volume in collaboration with the target frame. Additionally, to enhance the accuracy and resilience of the network structure, we introduce an attention-based depth net architecture to effectively integrate information from feature maps with varying resolutions. Compared to methods with similar computational costs, MGDepth achieves a significant reduction of approximately seven percent in root-mean-square error for self-supervised monocular depth estimation on the KITTI-2015 dataset.
A Compacted Structure for Cross-domain learning on Monocular Depth and Flow Estimation
Aug 25, 2022



Abstract:Accurate motion and depth recovery is important for many robot vision tasks including autonomous driving. Most previous studies have achieved cooperative multi-task interaction via either pre-defined loss functions or cross-domain prediction. This paper presents a multi-task scheme that achieves mutual assistance by means of our Flow to Depth (F2D), Depth to Flow (D2F), and Exponential Moving Average (EMA). F2D and D2F mechanisms enable multi-scale information integration between optical flow and depth domain based on differentiable shallow nets. A dual-head mechanism is used to predict optical flow for rigid and non-rigid motion based on a divide-and-conquer manner, which significantly improves the optical flow estimation performance. Furthermore, to make the prediction more robust and stable, EMA is used for our multi-task training. Experimental results on KITTI datasets show that our multi-task scheme outperforms other multi-task schemes and provide marked improvements on the prediction results.
Self-Supervised Depth Estimation in Laparoscopic Image using 3D Geometric Consistency
Aug 17, 2022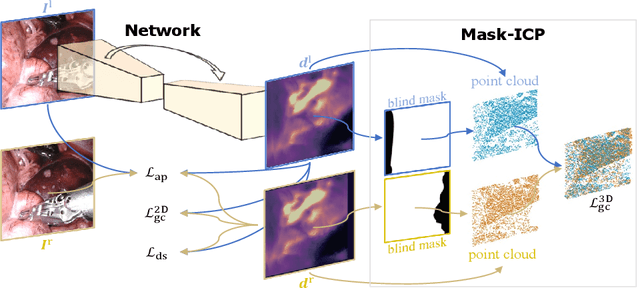
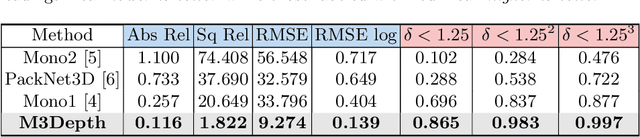
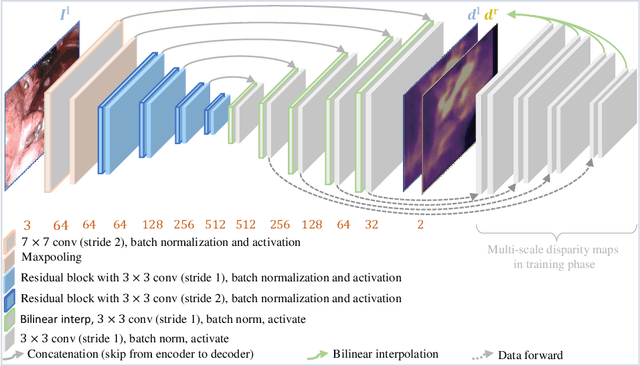
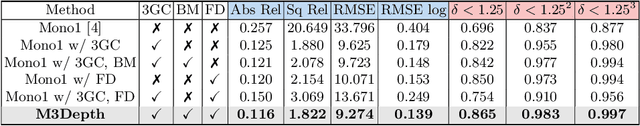
Abstract:Depth estimation is a crucial step for image-guided intervention in robotic surgery and laparoscopic imaging system. Since per-pixel depth ground truth is difficult to acquire for laparoscopic image data, it is rarely possible to apply supervised depth estimation to surgical applications. As an alternative, self-supervised methods have been introduced to train depth estimators using only synchronized stereo image pairs. However, most recent work focused on the left-right consistency in 2D and ignored valuable inherent 3D information on the object in real world coordinates, meaning that the left-right 3D geometric structural consistency is not fully utilized. To overcome this limitation, we present M3Depth, a self-supervised depth estimator to leverage 3D geometric structural information hidden in stereo pairs while keeping monocular inference. The method also removes the influence of border regions unseen in at least one of the stereo images via masking, to enhance the correspondences between left and right images in overlapping areas. Intensive experiments show that our method outperforms previous self-supervised approaches on both a public dataset and a newly acquired dataset by a large margin, indicating a good generalization across different samples and laparoscopes.
When CNN Meet with ViT: Towards Semi-Supervised Learning for Multi-Class Medical Image Semantic Segmentation
Aug 12, 2022
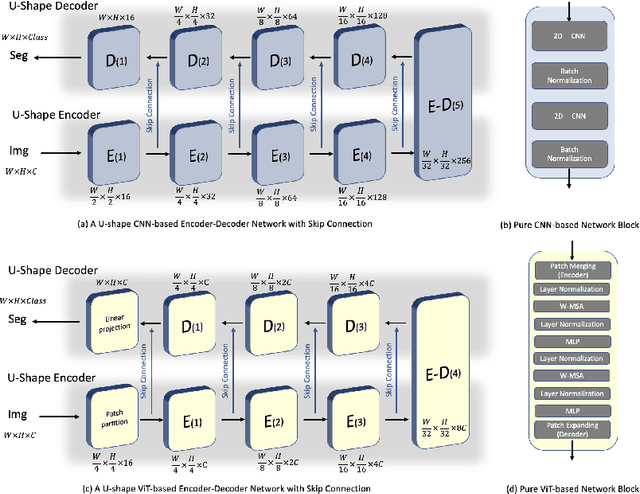


Abstract:Due to the lack of quality annotation in medical imaging community, semi-supervised learning methods are highly valued in image semantic segmentation tasks. In this paper, an advanced consistency-aware pseudo-label-based self-ensembling approach is presented to fully utilize the power of Vision Transformer(ViT) and Convolutional Neural Network(CNN) in semi-supervised learning. Our proposed framework consists of a feature-learning module which is enhanced by ViT and CNN mutually, and a guidance module which is robust for consistency-aware purposes. The pseudo labels are inferred and utilized recurrently and separately by views of CNN and ViT in the feature-learning module to expand the data set and are beneficial to each other. Meanwhile, a perturbation scheme is designed for the feature-learning module, and averaging network weight is utilized to develop the guidance module. By doing so, the framework combines the feature-learning strength of CNN and ViT, strengthens the performance via dual-view co-training, and enables consistency-aware supervision in a semi-supervised manner. A topological exploration of all alternative supervision modes with CNN and ViT are detailed validated, demonstrating the most promising performance and specific setting of our method on semi-supervised medical image segmentation tasks. Experimental results show that the proposed method achieves state-of-the-art performance on a public benchmark data set with a variety of metrics. The code is publicly available.
Recurrent Image Registration using Mutual Attention based Network
Jun 04, 2022



Abstract:Image registration is an important task in medical imaging which estimates the spatial transformation between different images. Many previous studies have used learning-based methods for multi-stage registration to perform 3D image registration to improve performance. The performance of the multi-stage approach, however, is limited by the size of the receptive field where complex motion does not occur at a single spatial scale. We propose a new registration network combining recursive network architecture and mutual attention mechanism to overcome these limitations. Compared with the previous deep learning methods, our network based on the recursive structure achieves the highest accuracy in lung Computed Tomography (CT) data set (Dice score of 92\% and average surface distance of 3.8mm for lungs) and one of the most accurate results in abdominal CT data set with 9 organs of various sizes (Dice score of 55\% and average surface distance of 7.8mm). We also showed that adding 3 recursive networks is sufficient to achieve the state-of-the-art results without a significant increase in the inference time.
Residual Aligner Network
Mar 07, 2022



Abstract:Image registration is important for medical imaging, the estimation of the spatial transformation between different images. Many previous studies have used learning-based methods for coarse-to-fine registration to efficiently perform 3D image registration. The coarse-to-fine approach, however, is limited when dealing with the different motions of nearby objects. Here we propose a novel Motion-Aware (MA) structure that captures the different motions in a region. The MA structure incorporates a novel Residual Aligner (RA) module which predicts the multi-head displacement field used to disentangle the different motions of multiple neighbouring objects. Compared with other deep learning methods, the network based on the MA structure and RA module achieve one of the most accurate unsupervised inter-subject registration on the 9 organs of assorted sizes in abdominal CT scans, with the highest-ranked registration of the veins (Dice Similarity Coefficient / Average surface distance: 62\%/4.9mm for the vena cava and 34\%/7.9mm for the portal and splenic vein), with a half-sized structure and more efficient computation. Applied to the segmentation of lungs in chest CT scans, the new network achieves results which were indistinguishable from the best-ranked networks (94\%/3.0mm). Additionally, the theorem on predicted motion pattern and the design of MA structure are validated by further analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge