Guang-Zhong Yang
Synergistic Development of Perovskite Memristors and Algorithms for Robust Analog Computing
Dec 03, 2024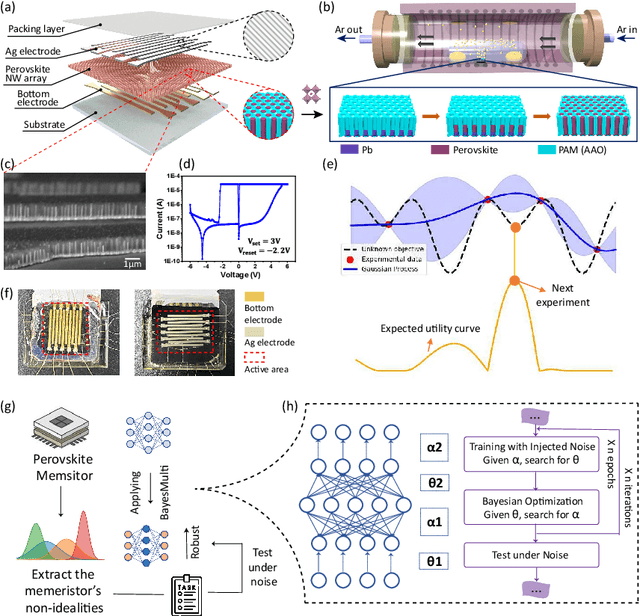
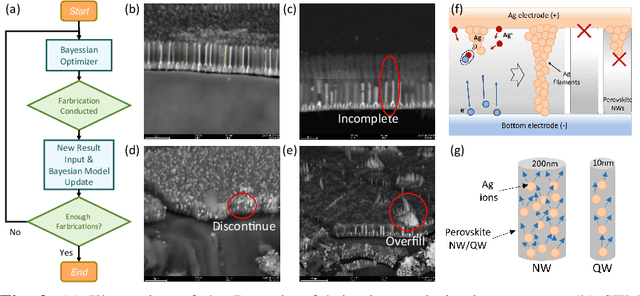
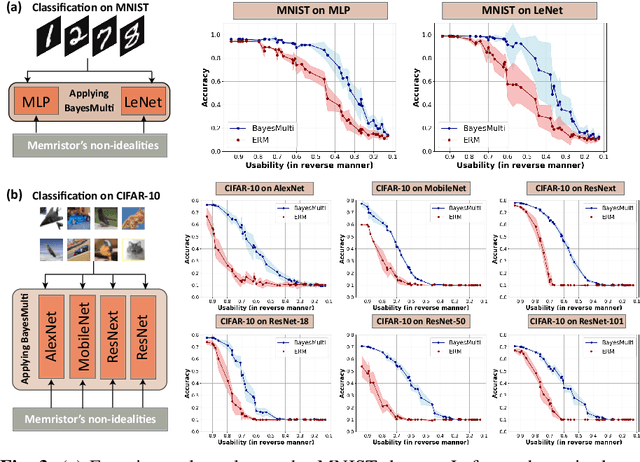
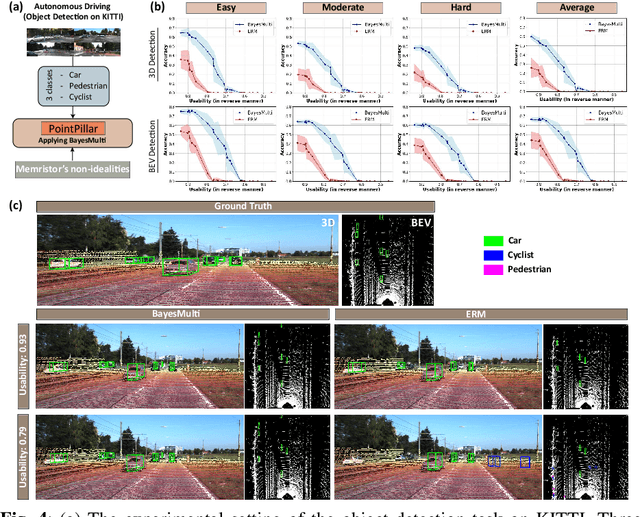
Abstract:Analog computing using non-volatile memristors has emerged as a promising solution for energy-efficient deep learning. New materials, like perovskites-based memristors are recently attractive due to their cost-effectiveness, energy efficiency and flexibility. Yet, challenges in material diversity and immature fabrications require extensive experimentation for device development. Moreover, significant non-idealities in these memristors often impede them for computing. Here, we propose a synergistic methodology to concurrently optimize perovskite memristor fabrication and develop robust analog DNNs that effectively address the inherent non-idealities of these memristors. Employing Bayesian optimization (BO) with a focus on usability, we efficiently identify optimal materials and fabrication conditions for perovskite memristors. Meanwhile, we developed "BayesMulti", a DNN training strategy utilizing BO-guided noise injection to improve the resistance of analog DNNs to memristor imperfections. Our approach theoretically ensures that within a certain range of parameter perturbations due to memristor non-idealities, the prediction outcomes remain consistent. Our integrated approach enables use of analog computing in much deeper and wider networks, which significantly outperforms existing methods in diverse tasks like image classification, autonomous driving, species identification, and large vision-language models, achieving up to 100-fold improvements. We further validate our methodology on a 10$\times$10 optimized perovskite memristor crossbar, demonstrating high accuracy in a classification task and low energy consumption. This study offers a versatile solution for efficient optimization of various analog computing systems, encompassing both devices and algorithms.
From Real Artifacts to Virtual Reference: A Robust Framework for Translating Endoscopic Images
Oct 23, 2024Abstract:Domain adaptation, which bridges the distributions across different modalities, plays a crucial role in multimodal medical image analysis. In endoscopic imaging, combining pre-operative data with intra-operative imaging is important for surgical planning and navigation. However, existing domain adaptation methods are hampered by distribution shift caused by in vivo artifacts, necessitating robust techniques for aligning noisy and artifact abundant patient endoscopic videos with clean virtual images reconstructed from pre-operative tomographic data for pose estimation during intraoperative guidance. This paper presents an artifact-resilient image translation method and an associated benchmark for this purpose. The method incorporates a novel ``local-global'' translation framework and a noise-resilient feature extraction strategy. For the former, it decouples the image translation process into a local step for feature denoising, and a global step for global style transfer. For feature extraction, a new contrastive learning strategy is proposed, which can extract noise-resilient features for establishing robust correspondence across domains. Detailed validation on both public and in-house clinical datasets has been conducted, demonstrating significantly improved performance compared to the current state-of-the-art.
LNQ 2023 challenge: Benchmark of weakly-supervised techniques for mediastinal lymph node quantification
Aug 19, 2024



Abstract:Accurate assessment of lymph node size in 3D CT scans is crucial for cancer staging, therapeutic management, and monitoring treatment response. Existing state-of-the-art segmentation frameworks in medical imaging often rely on fully annotated datasets. However, for lymph node segmentation, these datasets are typically small due to the extensive time and expertise required to annotate the numerous lymph nodes in 3D CT scans. Weakly-supervised learning, which leverages incomplete or noisy annotations, has recently gained interest in the medical imaging community as a potential solution. Despite the variety of weakly-supervised techniques proposed, most have been validated only on private datasets or small publicly available datasets. To address this limitation, the Mediastinal Lymph Node Quantification (LNQ) challenge was organized in conjunction with the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to advance weakly-supervised segmentation methods by providing a new, partially annotated dataset and a robust evaluation framework. A total of 16 teams from 5 countries submitted predictions to the validation leaderboard, and 6 teams from 3 countries participated in the evaluation phase. The results highlighted both the potential and the current limitations of weakly-supervised approaches. On one hand, weakly-supervised approaches obtained relatively good performance with a median Dice score of $61.0\%$. On the other hand, top-ranked teams, with a median Dice score exceeding $70\%$, boosted their performance by leveraging smaller but fully annotated datasets to combine weak supervision and full supervision. This highlights both the promise of weakly-supervised methods and the ongoing need for high-quality, fully annotated data to achieve higher segmentation performance.
Efficient Domain Adaptation for Endoscopic Visual Odometry
Mar 16, 2024



Abstract:Visual odometry plays a crucial role in endoscopic imaging, yet the scarcity of realistic images with ground truth poses poses a significant challenge. Therefore, domain adaptation offers a promising approach to bridge the pre-operative planning domain with the intra-operative real domain for learning odometry information. However, existing methodologies suffer from inefficiencies in the training time. In this work, an efficient neural style transfer framework for endoscopic visual odometry is proposed, which compresses the time from pre-operative planning to testing phase to less than five minutes. For efficient traing, this work focuses on training modules with only a limited number of real images and we exploit pre-operative prior information to dramatically reduce training duration. Moreover, during the testing phase, we propose a novel Test Time Adaptation (TTA) method to mitigate the gap in lighting conditions between training and testing datasets. Experimental evaluations conducted on two public endoscope datasets showcase that our method achieves state-of-the-art accuracy in visual odometry tasks while boasting the fastest training speeds. These results demonstrate significant promise for intra-operative surgery applications.
CDFI: Cross Domain Feature Interaction for Robust Bronchi Lumen Detection
Apr 18, 2023



Abstract:Endobronchial intervention is increasingly used as a minimally invasive means for the treatment of pulmonary diseases. In order to reduce the difficulty of manipulation in complex airway networks, robust lumen detection is essential for intraoperative guidance. However, these methods are sensitive to visual artifacts which are inevitable during the surgery. In this work, a cross domain feature interaction (CDFI) network is proposed to extract the structural features of lumens, as well as to provide artifact cues to characterize the visual features. To effectively extract the structural and artifact features, the Quadruple Feature Constraints (QFC) module is designed to constrain the intrinsic connections of samples with various imaging-quality. Furthermore, we design a Guided Feature Fusion (GFF) module to supervise the model for adaptive feature fusion based on different types of artifacts. Results show that the features extracted by the proposed method can preserve the structural information of lumen in the presence of large visual variations, bringing much-improved lumen detection accuracy.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
MR Elastography with Optimization-Based Phase Unwrapping and Traveling Wave Expansion-based Neural Network (TWENN)
Jan 06, 2023



Abstract:Magnetic Resonance Elastography (MRE) can characterize biomechanical properties of soft tissue for disease diagnosis and treatment planning. However, complicated wavefields acquired from MRE coupled with noise pose challenges for accurate displacement extraction and modulus estimation. Here we propose a pipeline for processing MRE images using optimization-based displacement extraction and Traveling Wave Expansion-based Neural Network (TWENN) modulus estimation. Phase unwrapping and displacement extraction were achieved by optimization of an objective function with Dual Data Consistency (Dual-DC). A complex-valued neural network using displacement covariance as input has been constructed for the estimation of complex wavenumbers. A model of traveling wave expansion is used to generate training datasets with different levels of noise for the network. The complex shear modulus map is obtained by a fusion of multifrequency and multidirectional data. Validation using images of brain and liver simulation demonstrates the practical value of the proposed pipeline, which can estimate the biomechanical properties with minimum root-mean-square-errors compared with state-of-the-art methods. Applications of the proposed method for processing MRE images of phantom, brain, and liver show clear anatomical features and that the pipeline is robust to noise and has a good generalization capability.
Revisiting Self-Supervised Contrastive Learning for Facial Expression Recognition
Oct 08, 2022
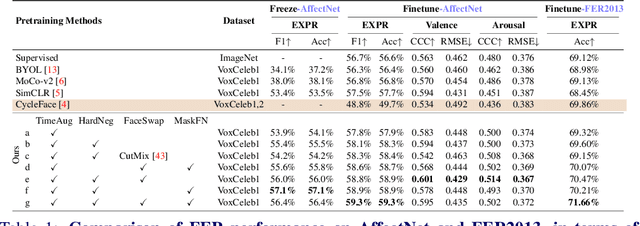
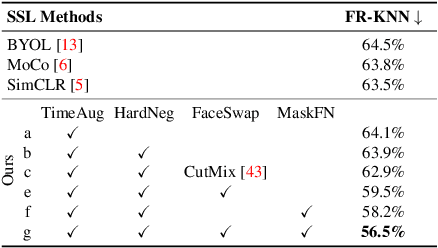

Abstract:The success of most advanced facial expression recognition works relies heavily on large-scale annotated datasets. However, it poses great challenges in acquiring clean and consistent annotations for facial expression datasets. On the other hand, self-supervised contrastive learning has gained great popularity due to its simple yet effective instance discrimination training strategy, which can potentially circumvent the annotation issue. Nevertheless, there remain inherent disadvantages of instance-level discrimination, which are even more challenging when faced with complicated facial representations. In this paper, we revisit the use of self-supervised contrastive learning and explore three core strategies to enforce expression-specific representations and to minimize the interference from other facial attributes, such as identity and face styling. Experimental results show that our proposed method outperforms the current state-of-the-art self-supervised learning methods, in terms of both categorical and dimensional facial expression recognition tasks.
Differentiable Topology-Preserved Distance Transform for Pulmonary Airway Segmentation
Sep 17, 2022



Abstract:Detailed pulmonary airway segmentation is a clinically important task for endobronchial intervention and treatment of peripheral lung cancer lesions. Convolutional Neural Networks (CNNs) are promising tools for medical image analysis but have been performing poorly for cases when there is a significantly imbalanced feature distribution, which is true for the airway data as the trachea and principal bronchi dominate most of the voxels whereas the lobar bronchi and distal segmental bronchi occupy only a small proportion. In this paper, we propose a Differentiable Topology-Preserved Distance Transform (DTPDT) framework to improve the performance of airway segmentation. A Topology-Preserved Surrogate (TPS) learning strategy is first proposed to equalize the training progress within-class distribution. Furthermore, a Convolutional Distance Transform (CDT) is designed to identify the breakage phenomenon with improved sensitivity, minimizing the variation of the distance map between the prediction and ground-truth. The proposed method is validated with the publicly available reference airway segmentation datasets.
A Compacted Structure for Cross-domain learning on Monocular Depth and Flow Estimation
Aug 25, 2022



Abstract:Accurate motion and depth recovery is important for many robot vision tasks including autonomous driving. Most previous studies have achieved cooperative multi-task interaction via either pre-defined loss functions or cross-domain prediction. This paper presents a multi-task scheme that achieves mutual assistance by means of our Flow to Depth (F2D), Depth to Flow (D2F), and Exponential Moving Average (EMA). F2D and D2F mechanisms enable multi-scale information integration between optical flow and depth domain based on differentiable shallow nets. A dual-head mechanism is used to predict optical flow for rigid and non-rigid motion based on a divide-and-conquer manner, which significantly improves the optical flow estimation performance. Furthermore, to make the prediction more robust and stable, EMA is used for our multi-task training. Experimental results on KITTI datasets show that our multi-task scheme outperforms other multi-task schemes and provide marked improvements on the prediction results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge