Yitong Zhang
The Mask of Civility: Benchmarking Chinese Mock Politeness Comprehension in Large Language Models
Feb 03, 2026Abstract:From a pragmatic perspective, this study systematically evaluates the differences in performance among representative large language models (LLMs) in recognizing politeness, impoliteness, and mock politeness phenomena in Chinese. Addressing the existing gaps in pragmatic comprehension, the research adopts the frameworks of Rapport Management Theory and the Model of Mock Politeness to construct a three-category dataset combining authentic and simulated Chinese discourse. Six representative models, including GPT-5.1 and DeepSeek, were selected as test subjects and evaluated under four prompting conditions: zero-shot, few-shot, knowledge-enhanced, and hybrid strategies. This study serves as a meaningful attempt within the paradigm of ``Great Linguistics,'' offering a novel approach to applying pragmatic theory in the age of technological transformation. It also responds to the contemporary question of how technology and the humanities may coexist, representing an interdisciplinary endeavor that bridges linguistic technology and humanistic reflection.
Lookahead-then-Verify: Reliable Constrained Decoding for Diffusion LLMs under Context-Free Grammars
Jan 31, 2026Abstract:Diffusion Large Language Models (dLLMs) have demonstrated promising generative capabilities and are increasingly used to produce formal languages defined by context-free grammars, such as source code and chemical expressions. However, as probabilistic models, they still struggle to generate syntactically valid outputs reliably. A natural and promising direction to address this issue is to adapt constrained decoding techniques to enforce grammatical correctness during generation. However, applying these techniques faces two primary obstacles. On the one hand, the non-autoregressive nature of dLLMs renders most existing constrained decoding approaches inapplicable. On the other hand, current approaches specifically designed for dLLMs may allow intermediate outputs that are impossible to complete into valid sentences, which significantly limits their reliability in practice. To address these challenges, we present LAVE, a constrained decoding approach specifically designed for dLLMs. Our approach leverages a key property of dLLMs, namely their ability to predict token distributions for all positions in parallel during each forward pass. Whenever a new token is proposed by model, LAVE performs lookahead using these distributions to efficiently and reliably verify the validity of the proposed token. This design ensures reliable constraints by reliably preserving the potential for intermediate outputs to be extended into valid sentences. Extensive experiments across four widely used dLLMs and three representative benchmarks demonstrate that LAVE consistently outperforms existing baselines and achieves substantial improvements in syntactic correctness, while incurring negligible runtime overhead.
AI-Driven Self-Evolving Software: A Promising Path Toward Software Automation
Oct 01, 2025Abstract:Software automation has long been a central goal of software engineering, striving for software development that proceeds without human intervention. Recent efforts have leveraged Artificial Intelligence (AI) to advance software automation with notable progress. However, current AI functions primarily as assistants to human developers, leaving software development still dependent on explicit human intervention. This raises a fundamental question: Can AI move beyond its role as an assistant to become a core component of software, thereby enabling genuine software automation? To investigate this vision, we introduce AI-Driven Self-Evolving Software, a new form of software that evolves continuously through direct interaction with users. We demonstrate the feasibility of this idea with a lightweight prototype built on a multi-agent architecture that autonomously interprets user requirements, generates and validates code, and integrates new functionalities. Case studies across multiple representative scenarios show that the prototype can reliably construct and reuse functionality, providing early evidence that such software systems can scale to more sophisticated applications and pave the way toward truly automated software development. We make code and cases in this work publicly available at https://anonymous.4open.science/r/live-software.
DAVSP: Safety Alignment for Large Vision-Language Models via Deep Aligned Visual Safety Prompt
Jun 11, 2025Abstract:Large Vision-Language Models (LVLMs) have achieved impressive progress across various applications but remain vulnerable to malicious queries that exploit the visual modality. Existing alignment approaches typically fail to resist malicious queries while preserving utility on benign ones effectively. To address these challenges, we propose Deep Aligned Visual Safety Prompt (DAVSP), which is built upon two key innovations. First, we introduce the Visual Safety Prompt, which appends a trainable padding region around the input image. It preserves visual features and expands the optimization space. Second, we propose Deep Alignment, a novel approach to train the visual safety prompt through supervision in the model's activation space. It enhances the inherent ability of LVLMs to perceive malicious queries, achieving deeper alignment than prior works. Extensive experiments across five benchmarks on two representative LVLMs demonstrate that DAVSP effectively resists malicious queries while preserving benign input utility. Furthermore, DAVSP exhibits great cross-model generation ability. Ablation studies further reveal that both the Visual Safety Prompt and Deep Alignment are essential components, jointly contributing to its overall effectiveness. The code is publicly available at https://github.com/zhangyitonggg/DAVSP.
Visual Adversarial Attack on Vision-Language Models for Autonomous Driving
Nov 27, 2024



Abstract:Vision-language models (VLMs) have significantly advanced autonomous driving (AD) by enhancing reasoning capabilities. However, these models remain highly vulnerable to adversarial attacks. While existing research has primarily focused on general VLM attacks, the development of attacks tailored to the safety-critical AD context has been largely overlooked. In this paper, we take the first step toward designing adversarial attacks specifically targeting VLMs in AD, exposing the substantial risks these attacks pose within this critical domain. We identify two unique challenges for effective adversarial attacks on AD VLMs: the variability of textual instructions and the time-series nature of visual scenarios. To this end, we propose ADvLM, the first visual adversarial attack framework specifically designed for VLMs in AD. Our framework introduces Semantic-Invariant Induction, which uses a large language model to create a diverse prompt library of textual instructions with consistent semantic content, guided by semantic entropy. Building on this, we introduce Scenario-Associated Enhancement, an approach where attention mechanisms select key frames and perspectives within driving scenarios to optimize adversarial perturbations that generalize across the entire scenario. Extensive experiments on several AD VLMs over multiple benchmarks show that ADvLM achieves state-of-the-art attack effectiveness. Moreover, real-world attack studies further validate its applicability and potential in practice.
DA-Code: Agent Data Science Code Generation Benchmark for Large Language Models
Oct 09, 2024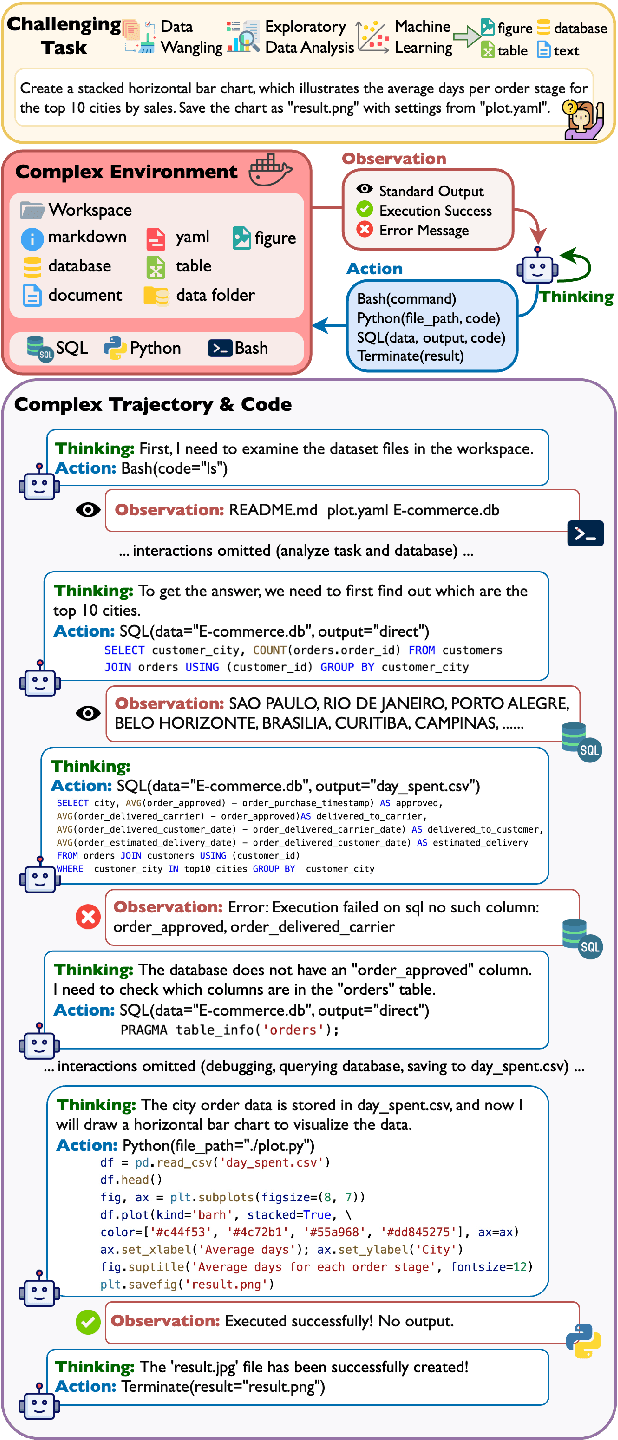
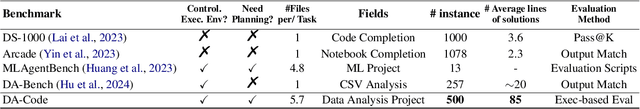
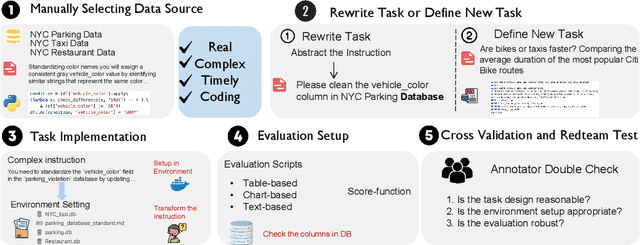
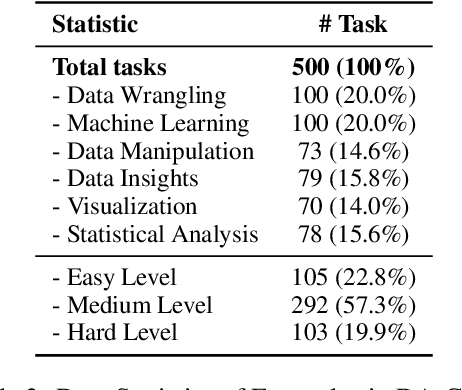
Abstract:We introduce DA-Code, a code generation benchmark specifically designed to assess LLMs on agent-based data science tasks. This benchmark features three core elements: First, the tasks within DA-Code are inherently challenging, setting them apart from traditional code generation tasks and demanding advanced coding skills in grounding and planning. Second, examples in DA-Code are all based on real and diverse data, covering a wide range of complex data wrangling and analytics tasks. Third, to solve the tasks, the models must utilize complex data science programming languages, to perform intricate data processing and derive the answers. We set up the benchmark in a controllable and executable environment that aligns with real-world data analysis scenarios and is scalable. The annotators meticulously design the evaluation suite to ensure the accuracy and robustness of the evaluation. We develop the DA-Agent baseline. Experiments show that although the baseline performs better than other existing frameworks, using the current best LLMs achieves only 30.5% accuracy, leaving ample room for improvement. We release our benchmark at [https://da-code-bench.github.io](https://da-code-bench.github.io).
PitVis-2023 Challenge: Workflow Recognition in videos of Endoscopic Pituitary Surgery
Sep 02, 2024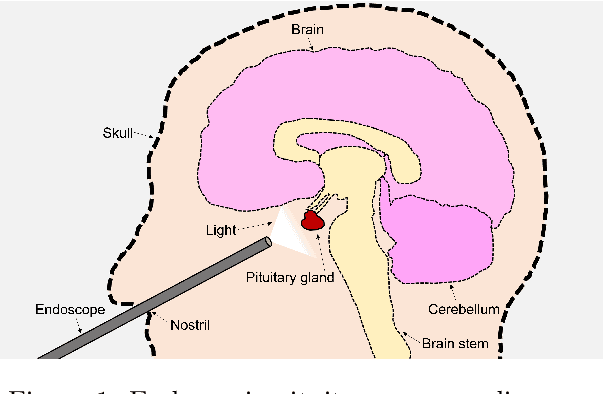
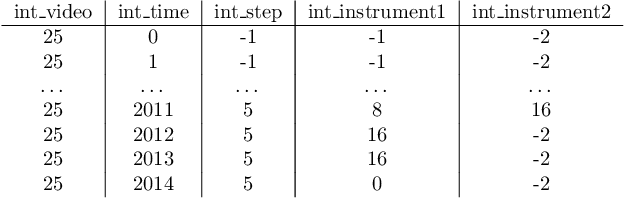
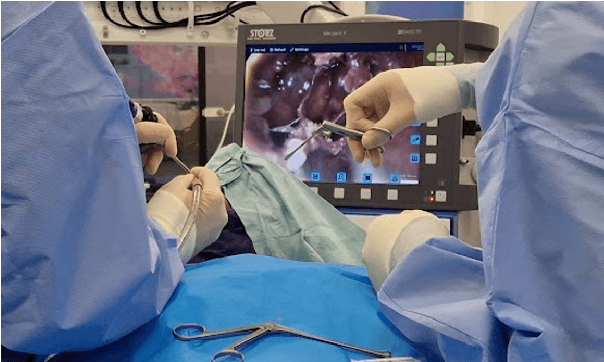
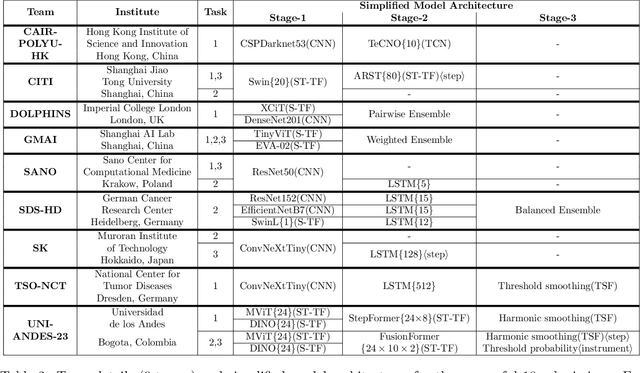
Abstract:The field of computer vision applied to videos of minimally invasive surgery is ever-growing. Workflow recognition pertains to the automated recognition of various aspects of a surgery: including which surgical steps are performed; and which surgical instruments are used. This information can later be used to assist clinicians when learning the surgery; during live surgery; and when writing operation notes. The Pituitary Vision (PitVis) 2023 Challenge tasks the community to step and instrument recognition in videos of endoscopic pituitary surgery. This is a unique task when compared to other minimally invasive surgeries due to the smaller working space, which limits and distorts vision; and higher frequency of instrument and step switching, which requires more precise model predictions. Participants were provided with 25-videos, with results presented at the MICCAI-2023 conference as part of the Endoscopic Vision 2023 Challenge in Vancouver, Canada, on 08-Oct-2023. There were 18-submissions from 9-teams across 6-countries, using a variety of deep learning models. A commonality between the top performing models was incorporating spatio-temporal and multi-task methods, with greater than 50% and 10% macro-F1-score improvement over purely spacial single-task models in step and instrument recognition respectively. The PitVis-2023 Challenge therefore demonstrates state-of-the-art computer vision models in minimally invasive surgery are transferable to a new dataset, with surgery specific techniques used to enhance performance, progressing the field further. Benchmark results are provided in the paper, and the dataset is publicly available at: https://doi.org/10.5522/04/26531686.
MonoPCC: Photometric-invariant Cycle Constraint for Monocular Depth Estimation of Endoscopic Images
Apr 25, 2024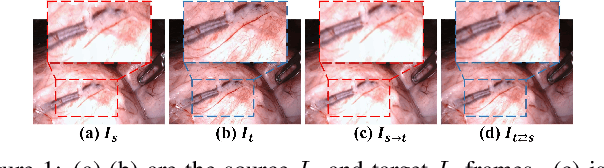
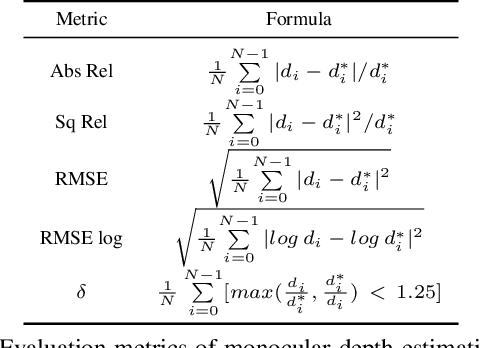
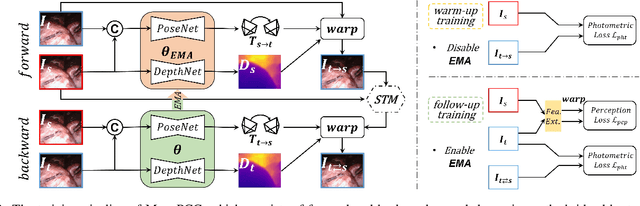
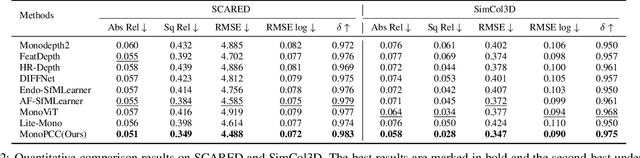
Abstract:Photometric constraint is indispensable for self-supervised monocular depth estimation. It involves warping a source image onto a target view using estimated depth&pose, and then minimizing the difference between the warped and target images. However, the endoscopic built-in light causes significant brightness fluctuations, and thus makes the photometric constraint unreliable. Previous efforts only mitigate this relying on extra models to calibrate image brightness. In this paper, we propose MonoPCC to address the brightness inconsistency radically by reshaping the photometric constraint into a cycle form. Instead of only warping the source image, MonoPCC constructs a closed loop consisting of two opposite forward-backward warping paths: from target to source and then back to target. Thus, the target image finally receives an image cycle-warped from itself, which naturally makes the constraint invariant to brightness changes. Moreover, MonoPCC transplants the source image's phase-frequency into the intermediate warped image to avoid structure lost, and also stabilizes the training via an exponential moving average (EMA) strategy to avoid frequent changes in the forward warping. The comprehensive and extensive experimental results on three datasets demonstrate that our proposed MonoPCC shows a great robustness to the brightness inconsistency, and exceeds other state-of-the-arts by reducing the absolute relative error by at least 7.27%.
IBoxCLA: Towards Robust Box-supervised Segmentation of Polyp via Improved Box-dice and Contrastive Latent-anchors
Oct 14, 2023



Abstract:Box-supervised polyp segmentation attracts increasing attention for its cost-effective potential. Existing solutions often rely on learning-free methods or pretrained models to laboriously generate pseudo masks, triggering Dice constraint subsequently. In this paper, we found that a model guided by the simplest box-filled masks can accurately predict polyp locations/sizes, but suffers from shape collapsing. In response, we propose two innovative learning fashions, Improved Box-dice (IBox) and Contrastive Latent-Anchors (CLA), and combine them to train a robust box-supervised model IBoxCLA. The core idea behind IBoxCLA is to decouple the learning of location/size and shape, allowing for focused constraints on each of them. Specifically, IBox transforms the segmentation map into a proxy map using shape decoupling and confusion-region swapping sequentially. Within the proxy map, shapes are disentangled, while locations/sizes are encoded as box-like responses. By constraining the proxy map instead of the raw prediction, the box-filled mask can well supervise IBoxCLA without misleading its shape learning. Furthermore, CLA contributes to shape learning by generating two types of latent anchors, which are learned and updated using momentum and segmented polyps to steadily represent polyp and background features. The latent anchors facilitate IBoxCLA to capture discriminative features within and outside boxes in a contrastive manner, yielding clearer boundaries. We benchmark IBoxCLA on five public polyp datasets. The experimental results demonstrate the competitive performance of IBoxCLA compared to recent fully-supervised polyp segmentation methods, and its superiority over other box-supervised state-of-the-arts with a relative increase of overall mDice and mIoU by at least 6.5% and 7.5%, respectively.
SurgT challenge: Benchmark of Soft-Tissue Trackers for Robotic Surgery
Feb 28, 2023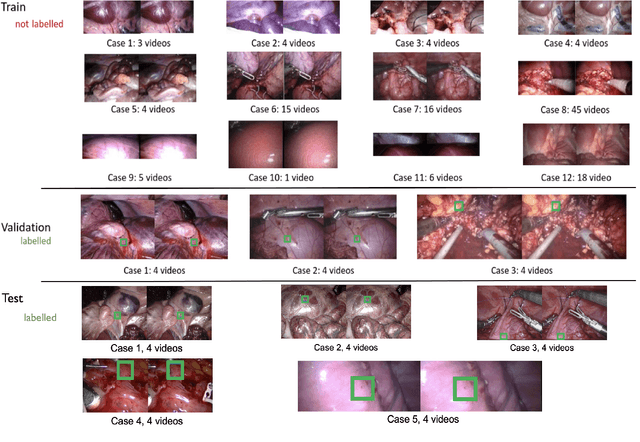
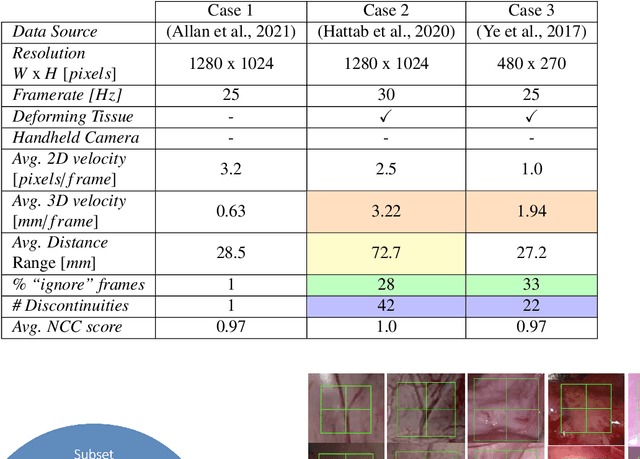
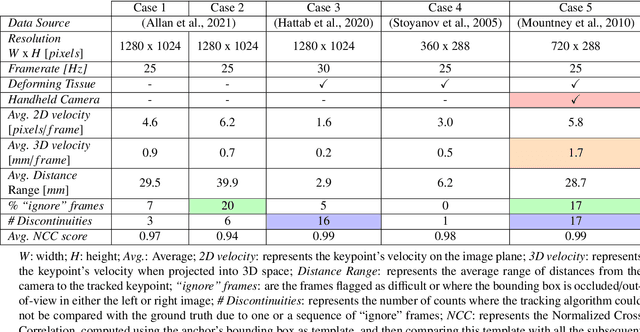
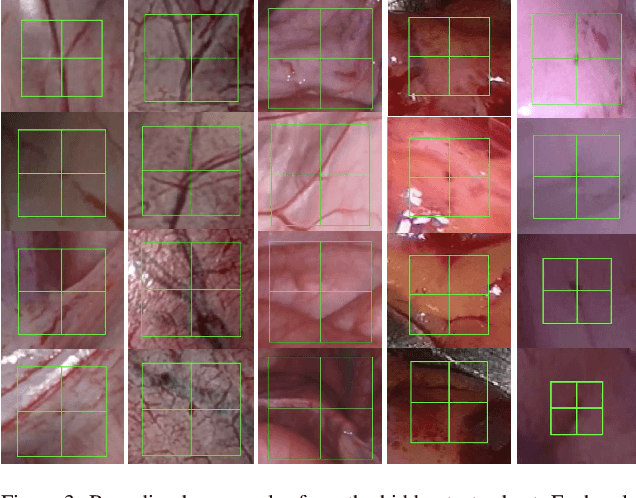
Abstract:This paper introduces the "SurgT: Surgical Tracking" challenge which was organised in conjunction with the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2022). There were two purposes for the creation of this challenge: (1) the establishment of the first standardised benchmark for the research community to assess soft-tissue trackers; and (2) to encourage the development of unsupervised deep learning methods, given the lack of annotated data in surgery. A dataset of 157 stereo endoscopic videos from 20 clinical cases, along with stereo camera calibration parameters, have been provided. The participants were tasked with the development of algorithms to track a bounding box on stereo endoscopic videos. At the end of the challenge, the developed methods were assessed on a previously hidden test subset. This assessment uses benchmarking metrics that were purposely developed for this challenge and are now available online. The teams were ranked according to their Expected Average Overlap (EAO) score, which is a weighted average of the Intersection over Union (IoU) scores. The performance evaluation study verifies the efficacy of unsupervised deep learning algorithms in tracking soft-tissue. The best-performing method achieved an EAO score of 0.583 in the test subset. The dataset and benchmarking tool created for this challenge have been made publicly available. This challenge is expected to contribute to the development of autonomous robotic surgery and other digital surgical technologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge