Alistair Weld
Confidence-Based Annotation Of Brain Tumours In Ultrasound
Feb 21, 2025



Abstract:Purpose: An investigation of the challenge of annotating discrete segmentations of brain tumours in ultrasound, with a focus on the issue of aleatoric uncertainty along the tumour margin, particularly for diffuse tumours. A segmentation protocol and method is proposed that incorporates this margin-related uncertainty while minimising the interobserver variance through reduced subjectivity, thereby diminishing annotator epistemic uncertainty. Approach: A sparse confidence method for annotation is proposed, based on a protocol designed using computer vision and radiology theory. Results: Output annotations using the proposed method are compared with the corresponding professional discrete annotation variance between the observers. A linear relationship was measured within the tumour margin region, with a Pearson correlation of 0.8. The downstream application was explored, comparing training using confidence annotations as soft labels with using the best discrete annotations as hard labels. In all evaluation folds, the Brier score was superior for the soft-label trained network. Conclusion: A formal framework was constructed to demonstrate the infeasibility of discrete annotation of brain tumours in B-mode ultrasound. Subsequently, a method for sparse confidence-based annotation is proposed and evaluated. Keywords: Brain tumours, ultrasound, confidence, annotation.
Standardisation of Convex Ultrasound Data Through Geometric Analysis and Augmentation
Feb 13, 2025



Abstract:The application of ultrasound in healthcare has seen increased diversity and importance. Unlike other medical imaging modalities, ultrasound research and development has historically lagged, particularly in the case of applications with data-driven algorithms. A significant issue with ultrasound is the extreme variability of the images, due to the number of different machines available and the possible combination of parameter settings. One outcome of this is the lack of standardised and benchmarking ultrasound datasets. The method proposed in this article is an approach to alleviating this issue of disorganisation. For this purpose, the issue of ultrasound data sparsity is examined and a novel perspective, approach, and solution is proposed; involving the extraction of the underlying ultrasound plane within the image and representing it using annulus sector geometry. An application of this methodology is proposed, which is the extraction of scan lines and the linearisation of convex planes. Validation of the robustness of the proposed method is performed on both private and public data. The impact of deformation and the invertibility of augmentation using the estimated annulus sector parameters is also studied. Keywords: Ultrasound, Annulus Sector, Augmentation, Linearisation.
SurgRIPE challenge: Benchmark of Surgical Robot Instrument Pose Estimation
Jan 06, 2025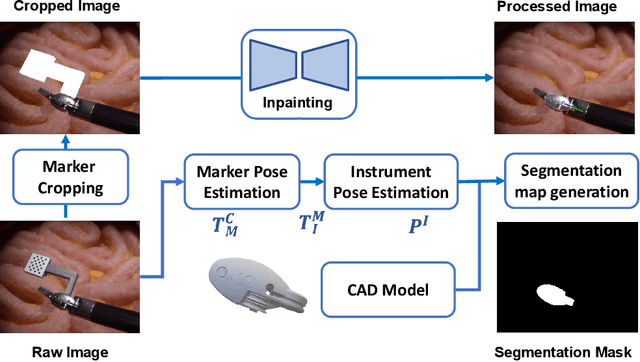

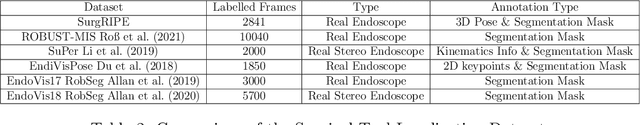
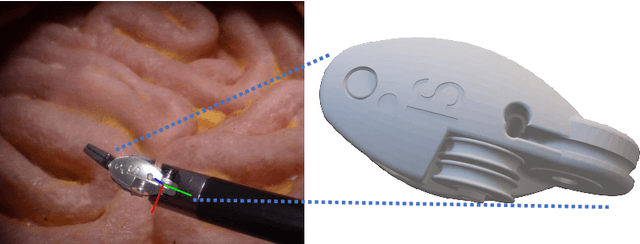
Abstract:Accurate instrument pose estimation is a crucial step towards the future of robotic surgery, enabling applications such as autonomous surgical task execution. Vision-based methods for surgical instrument pose estimation provide a practical approach to tool tracking, but they often require markers to be attached to the instruments. Recently, more research has focused on the development of marker-less methods based on deep learning. However, acquiring realistic surgical data, with ground truth instrument poses, required for deep learning training, is challenging. To address the issues in surgical instrument pose estimation, we introduce the Surgical Robot Instrument Pose Estimation (SurgRIPE) challenge, hosted at the 26th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2023. The objectives of this challenge are: (1) to provide the surgical vision community with realistic surgical video data paired with ground truth instrument poses, and (2) to establish a benchmark for evaluating markerless pose estimation methods. The challenge led to the development of several novel algorithms that showcased improved accuracy and robustness over existing methods. The performance evaluation study on the SurgRIPE dataset highlights the potential of these advanced algorithms to be integrated into robotic surgery systems, paving the way for more precise and autonomous surgical procedures. The SurgRIPE challenge has successfully established a new benchmark for the field, encouraging further research and development in surgical robot instrument pose estimation.
Towards Safe and Collaborative Robotic Ultrasound Tissue Scanning in Neurosurgery
Jan 04, 2024



Abstract:Intraoperative ultrasound imaging is used to facilitate safe brain tumour resection. However, due to challenges with image interpretation and the physical scanning, this tool has yet to achieve widespread adoption in neurosurgery. In this paper, we introduce the components and workflow of a novel, versatile robotic platform for intraoperative ultrasound tissue scanning in neurosurgery. An RGB-D camera attached to the robotic arm allows for automatic object localisation with ArUco markers, and 3D surface reconstruction as a triangular mesh using the ImFusion Suite software solution. Impedance controlled guidance of the US probe along arbitrary surfaces, represented as a mesh, enables collaborative US scanning, i.e., autonomous, teleoperated and hands-on guided data acquisition. A preliminary experiment evaluates the suitability of the conceptual workflow and system components for probe landing on a custom-made soft-tissue phantom. Further assessment in future experiments will be necessary to prove the effectiveness of the presented platform.
Identifying Visible Tissue in Intraoperative Ultrasound Images during Brain Surgery: A Method and Application
Jun 01, 2023



Abstract:Intraoperative ultrasound scanning is a demanding visuotactile task. It requires operators to simultaneously localise the ultrasound perspective and manually perform slight adjustments to the pose of the probe, making sure not to apply excessive force or breaking contact with the tissue, whilst also characterising the visible tissue. In this paper, we propose a method for the identification of the visible tissue, which enables the analysis of ultrasound probe and tissue contact via the detection of acoustic shadow and construction of confidence maps of the perceptual salience. Detailed validation with both in vivo and phantom data is performed. First, we show that our technique is capable of achieving state of the art acoustic shadow scan line classification - with an average binary classification accuracy on unseen data of 0.87. Second, we show that our framework for constructing confidence maps is able to produce an ideal response to a probe's pose that is being oriented in and out of optimality - achieving an average RMSE across five scans of 0.174. The performance evaluation justifies the potential clinical value of the method which can be used both to assist clinical training and optimise robot-assisted ultrasound tissue scanning.
Automated robotic intraoperative ultrasound for brain surgery
Apr 03, 2023


Abstract:During brain tumour resection, localising cancerous tissue and delineating healthy and pathological borders is challenging, even for experienced neurosurgeons and neuroradiologists. Intraoperative imaging is commonly employed for determining and updating surgical plans in the operating room. Ultrasound (US) has presented itself a suitable tool for this task, owing to its ease of integration into the operating room and surgical procedure. However, widespread establishment of this tool has been limited because of the difficulty of anatomy localisation and data interpretation. In this work, we present a robotic framework designed and tested on a soft-tissue-mimicking brain phantom, simulating intraoperative US (iUS) scanning during brain tumour surgery.
SurgT challenge: Benchmark of Soft-Tissue Trackers for Robotic Surgery
Feb 28, 2023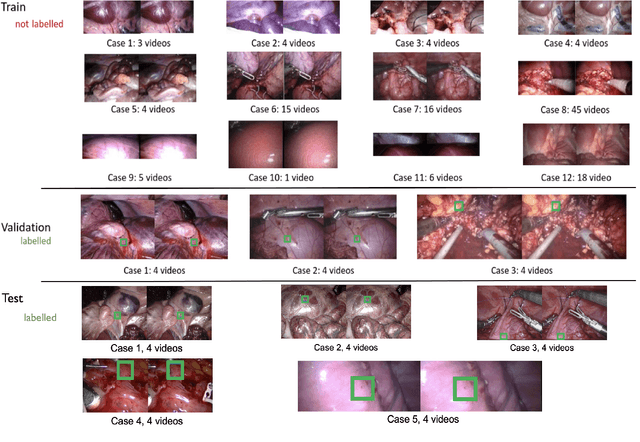
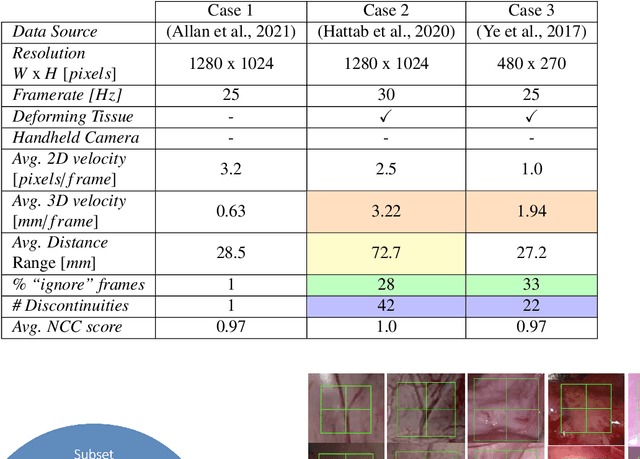
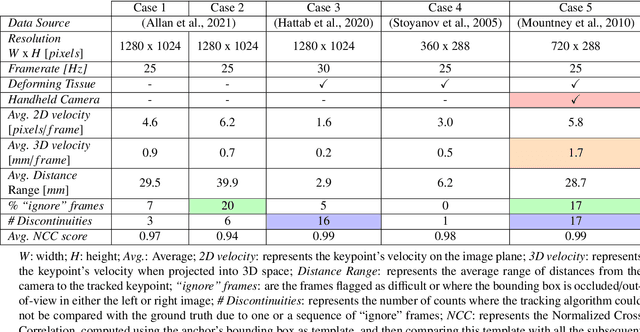
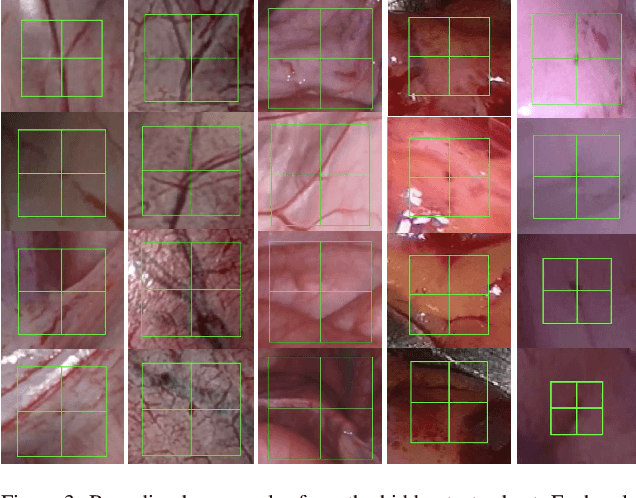
Abstract:This paper introduces the "SurgT: Surgical Tracking" challenge which was organised in conjunction with the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2022). There were two purposes for the creation of this challenge: (1) the establishment of the first standardised benchmark for the research community to assess soft-tissue trackers; and (2) to encourage the development of unsupervised deep learning methods, given the lack of annotated data in surgery. A dataset of 157 stereo endoscopic videos from 20 clinical cases, along with stereo camera calibration parameters, have been provided. The participants were tasked with the development of algorithms to track a bounding box on stereo endoscopic videos. At the end of the challenge, the developed methods were assessed on a previously hidden test subset. This assessment uses benchmarking metrics that were purposely developed for this challenge and are now available online. The teams were ranked according to their Expected Average Overlap (EAO) score, which is a weighted average of the Intersection over Union (IoU) scores. The performance evaluation study verifies the efficacy of unsupervised deep learning algorithms in tracking soft-tissue. The best-performing method achieved an EAO score of 0.583 in the test subset. The dataset and benchmarking tool created for this challenge have been made publicly available. This challenge is expected to contribute to the development of autonomous robotic surgery and other digital surgical technologies.
Regularizing disparity estimation via multi task learning with structured light reconstruction
Jan 19, 2023Abstract:3D reconstruction is a useful tool for surgical planning and guidance. However, the lack of available medical data stunts research and development in this field, as supervised deep learning methods for accurate disparity estimation rely heavily on large datasets containing ground truth information. Alternative approaches to supervision have been explored, such as self-supervision, which can reduce or remove entirely the need for ground truth. However, no proposed alternatives have demonstrated performance capabilities close to what would be expected from a supervised setup. This work aims to alleviate this issue. In this paper, we investigate the learning of structured light projections to enhance the development of direct disparity estimation networks. We show for the first time that it is possible to accurately learn the projection of structured light on a scene, implicitly learning disparity. Secondly, we \textcolor{black}{explore the use of a multi task learning (MTL) framework for the joint training of structured light and disparity. We present results which show that MTL with structured light improves disparity training; without increasing the number of model parameters. Our MTL setup outperformed the single task learning (STL) network in every validation test. Notably, in the medical generalisation test, the STL error was 1.4 times worse than that of the best MTL performance. The benefit of using MTL is emphasised when the training data is limited.} A dataset containing stereoscopic images, disparity maps and structured light projections on medical phantoms and ex vivo tissue was created for evaluation together with virtual scenes. This dataset will be made publicly available in the future.
Collaborative Robotic Ultrasound Tissue Scanning for Surgical Resection Guidance in Neurosurgery
Jan 19, 2023

Abstract:The aim of this paper is to introduce a robotic platform for autonomous iUS tissue scanning to optimise intraoperative diagnosis and improve surgical resection during robot-assisted operations. To guide anatomy specific robotic scanning and generate a representation of the robot task space, fast and accurate techniques for the recovery of 3D morphological structures of the surgical cavity are developed. The prototypic DLR MIRO surgical robotic arm is used to control the applied force and the in-plane motion of the US transducer. A key application of the proposed platform is the scanning of brain tissue to guide tumour resection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge