Mert Asim Karaoglu
UnReflectAnything: RGB-Only Highlight Removal by Rendering Synthetic Specular Supervision
Dec 11, 2025Abstract:Specular highlights distort appearance, obscure texture, and hinder geometric reasoning in both natural and surgical imagery. We present UnReflectAnything, an RGB-only framework that removes highlights from a single image by predicting a highlight map together with a reflection-free diffuse reconstruction. The model uses a frozen vision transformer encoder to extract multi-scale features, a lightweight head to localize specular regions, and a token-level inpainting module that restores corrupted feature patches before producing the final diffuse image. To overcome the lack of paired supervision, we introduce a Virtual Highlight Synthesis pipeline that renders physically plausible specularities using monocular geometry, Fresnel-aware shading, and randomized lighting which enables training on arbitrary RGB images with correct geometric structure. UnReflectAnything generalizes across natural and surgical domains where non-Lambertian surfaces and non-uniform lighting create severe highlights and it achieves competitive performance with state-of-the-art results on several benchmarks. Project Page: https://alberto-rota.github.io/UnReflectAnything/
LiteTracker: Leveraging Temporal Causality for Accurate Low-latency Tissue Tracking
Apr 14, 2025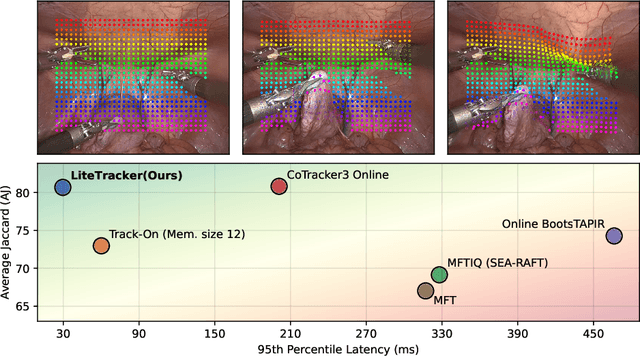
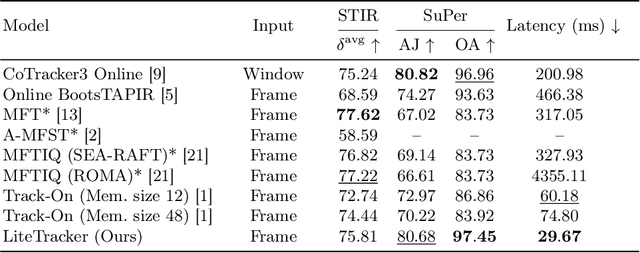
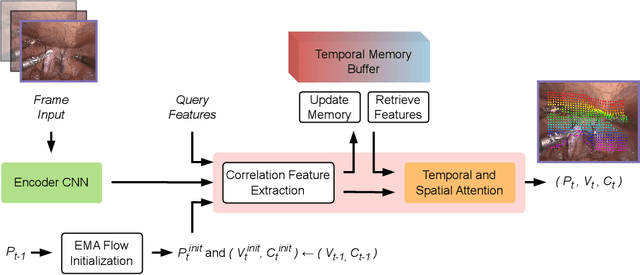
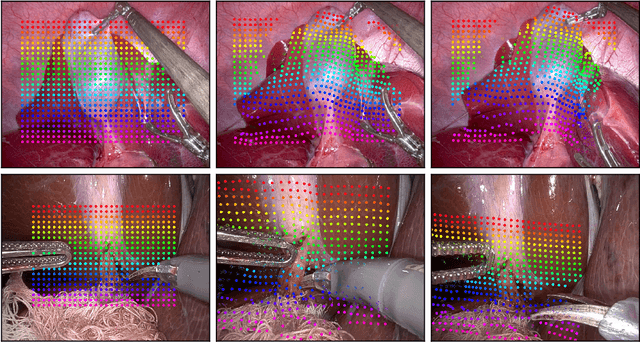
Abstract:Tissue tracking plays a critical role in various surgical navigation and extended reality (XR) applications. While current methods trained on large synthetic datasets achieve high tracking accuracy and generalize well to endoscopic scenes, their runtime performances fail to meet the low-latency requirements necessary for real-time surgical applications. To address this limitation, we propose LiteTracker, a low-latency method for tissue tracking in endoscopic video streams. LiteTracker builds on a state-of-the-art long-term point tracking method, and introduces a set of training-free runtime optimizations. These optimizations enable online, frame-by-frame tracking by leveraging a temporal memory buffer for efficient feature reuse and utilizing prior motion for accurate track initialization. LiteTracker demonstrates significant runtime improvements being around 7x faster than its predecessor and 2x than the state-of-the-art. Beyond its primary focus on efficiency, LiteTracker delivers high-accuracy tracking and occlusion prediction, performing competitively on both the STIR and SuPer datasets. We believe LiteTracker is an important step toward low-latency tissue tracking for real-time surgical applications in the operating room.
Point Tracking in Surgery--The 2024 Surgical Tattoos in Infrared (STIR) Challenge
Mar 31, 2025Abstract:Understanding tissue motion in surgery is crucial to enable applications in downstream tasks such as segmentation, 3D reconstruction, virtual tissue landmarking, autonomous probe-based scanning, and subtask autonomy. Labeled data are essential to enabling algorithms in these downstream tasks since they allow us to quantify and train algorithms. This paper introduces a point tracking challenge to address this, wherein participants can submit their algorithms for quantification. The submitted algorithms are evaluated using a dataset named surgical tattoos in infrared (STIR), with the challenge aptly named the STIR Challenge 2024. The STIR Challenge 2024 comprises two quantitative components: accuracy and efficiency. The accuracy component tests the accuracy of algorithms on in vivo and ex vivo sequences. The efficiency component tests the latency of algorithm inference. The challenge was conducted as a part of MICCAI EndoVis 2024. In this challenge, we had 8 total teams, with 4 teams submitting before and 4 submitting after challenge day. This paper details the STIR Challenge 2024, which serves to move the field towards more accurate and efficient algorithms for spatial understanding in surgery. In this paper we summarize the design, submissions, and results from the challenge. The challenge dataset is available here: https://zenodo.org/records/14803158 , and the code for baseline models and metric calculation is available here: https://github.com/athaddius/STIRMetrics
SurgRIPE challenge: Benchmark of Surgical Robot Instrument Pose Estimation
Jan 06, 2025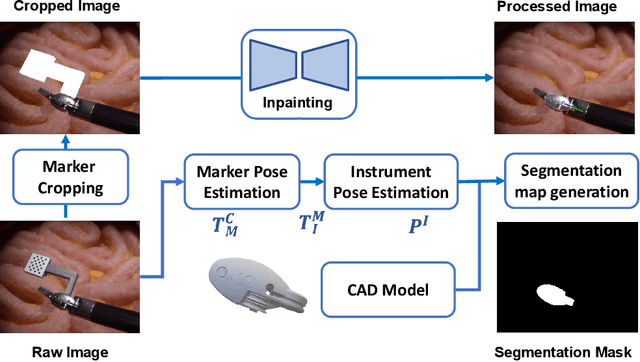

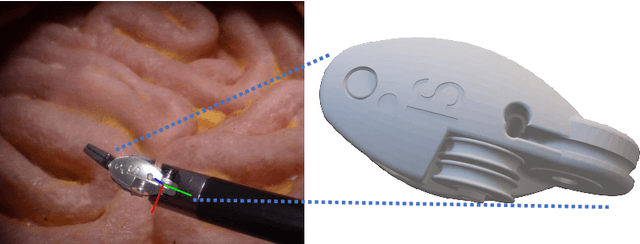
Abstract:Accurate instrument pose estimation is a crucial step towards the future of robotic surgery, enabling applications such as autonomous surgical task execution. Vision-based methods for surgical instrument pose estimation provide a practical approach to tool tracking, but they often require markers to be attached to the instruments. Recently, more research has focused on the development of marker-less methods based on deep learning. However, acquiring realistic surgical data, with ground truth instrument poses, required for deep learning training, is challenging. To address the issues in surgical instrument pose estimation, we introduce the Surgical Robot Instrument Pose Estimation (SurgRIPE) challenge, hosted at the 26th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2023. The objectives of this challenge are: (1) to provide the surgical vision community with realistic surgical video data paired with ground truth instrument poses, and (2) to establish a benchmark for evaluating markerless pose estimation methods. The challenge led to the development of several novel algorithms that showcased improved accuracy and robustness over existing methods. The performance evaluation study on the SurgRIPE dataset highlights the potential of these advanced algorithms to be integrated into robotic surgery systems, paving the way for more precise and autonomous surgical procedures. The SurgRIPE challenge has successfully established a new benchmark for the field, encouraging further research and development in surgical robot instrument pose estimation.
FLex: Joint Pose and Dynamic Radiance Fields Optimization for Stereo Endoscopic Videos
Mar 18, 2024Abstract:Reconstruction of endoscopic scenes is an important asset for various medical applications, from post-surgery analysis to educational training. Neural rendering has recently shown promising results in endoscopic reconstruction with deforming tissue. However, the setup has been restricted to a static endoscope, limited deformation, or required an external tracking device to retrieve camera pose information of the endoscopic camera. With FLex we adress the challenging setup of a moving endoscope within a highly dynamic environment of deforming tissue. We propose an implicit scene separation into multiple overlapping 4D neural radiance fields (NeRFs) and a progressive optimization scheme jointly optimizing for reconstruction and camera poses from scratch. This improves the ease-of-use and allows to scale reconstruction capabilities in time to process surgical videos of 5,000 frames and more; an improvement of more than ten times compared to the state of the art while being agnostic to external tracking information. Extensive evaluations on the StereoMIS dataset show that FLex significantly improves the quality of novel view synthesis while maintaining competitive pose accuracy.
RIDE: Self-Supervised Learning of Rotation-Equivariant Keypoint Detection and Invariant Description for Endoscopy
Sep 18, 2023



Abstract:Unlike in natural images, in endoscopy there is no clear notion of an up-right camera orientation. Endoscopic videos therefore often contain large rotational motions, which require keypoint detection and description algorithms to be robust to these conditions. While most classical methods achieve rotation-equivariant detection and invariant description by design, many learning-based approaches learn to be robust only up to a certain degree. At the same time learning-based methods under moderate rotations often outperform classical approaches. In order to address this shortcoming, in this paper we propose RIDE, a learning-based method for rotation-equivariant detection and invariant description. Following recent advancements in group-equivariant learning, RIDE models rotation-equivariance implicitly within its architecture. Trained in a self-supervised manner on a large curation of endoscopic images, RIDE requires no manual labeling of training data. We test RIDE in the context of surgical tissue tracking on the SuPeR dataset as well as in the context of relative pose estimation on a repurposed version of the SCARED dataset. In addition we perform explicit studies showing its robustness to large rotations. Our comparison against recent learning-based and classical approaches shows that RIDE sets a new state-of-the-art performance on matching and relative pose estimation tasks and scores competitively on surgical tissue tracking.
DynaMoN: Motion-Aware Fast And Robust Camera Localization for Dynamic NeRF
Sep 16, 2023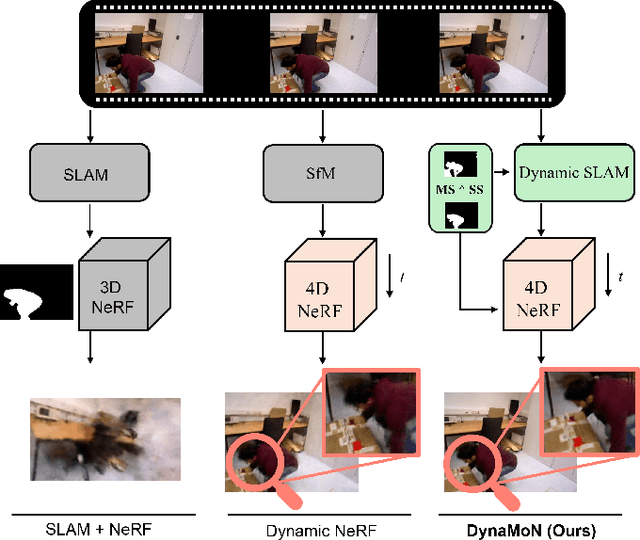

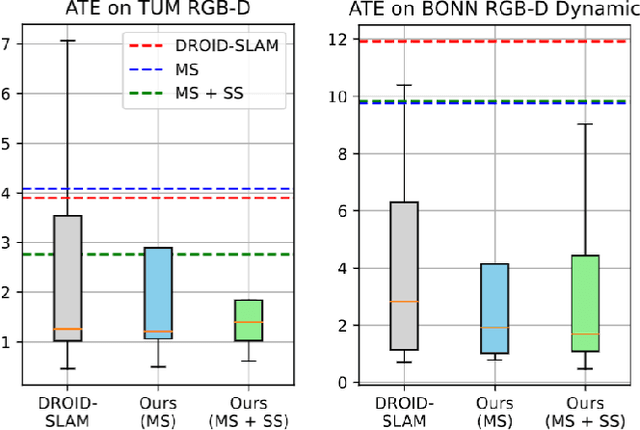

Abstract:Dynamic reconstruction with neural radiance fields (NeRF) requires accurate camera poses. These are often hard to retrieve with existing structure-from-motion (SfM) pipelines as both camera and scene content can change. We propose DynaMoN that leverages simultaneous localization and mapping (SLAM) jointly with motion masking to handle dynamic scene content. Our robust SLAM-based tracking module significantly accelerates the training process of the dynamic NeRF while improving the quality of synthesized views at the same time. Extensive experimental validation on TUM RGB-D, BONN RGB-D Dynamic and the DyCheck's iPhone dataset, three real-world datasets, shows the advantages of DynaMoN both for camera pose estimation and novel view synthesis.
Adversarial Domain Feature Adaptation for Bronchoscopic Depth Estimation
Sep 24, 2021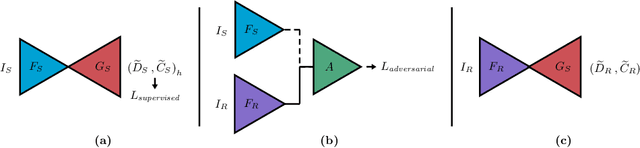
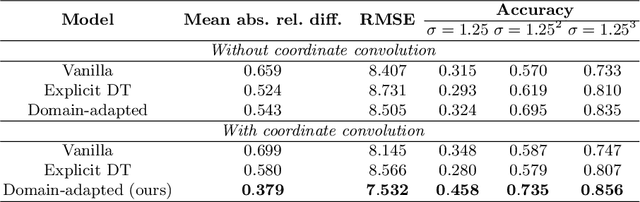
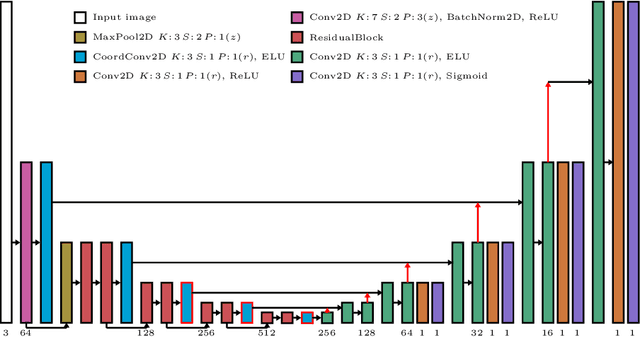
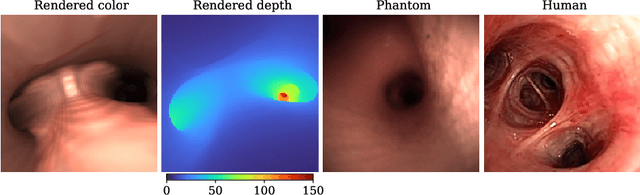
Abstract:Depth estimation from monocular images is an important task in localization and 3D reconstruction pipelines for bronchoscopic navigation. Various supervised and self-supervised deep learning-based approaches have proven themselves on this task for natural images. However, the lack of labeled data and the bronchial tissue's feature-scarce texture make the utilization of these methods ineffective on bronchoscopic scenes. In this work, we propose an alternative domain-adaptive approach. Our novel two-step structure first trains a depth estimation network with labeled synthetic images in a supervised manner; then adopts an unsupervised adversarial domain feature adaptation scheme to improve the performance on real images. The results of our experiments show that the proposed method improves the network's performance on real images by a considerable margin and can be employed in 3D reconstruction pipelines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge