Ihsan Ullah
TAU, LISN
Stylized Meta-Album: Group-bias injection with style transfer to study robustness against distribution shifts
Dec 10, 2025Abstract:We introduce Stylized Meta-Album (SMA), a new image classification meta-dataset comprising 24 datasets (12 content datasets, and 12 stylized datasets), designed to advance studies on out-of-distribution (OOD) generalization and related topics. Created using style transfer techniques from 12 subject classification datasets, SMA provides a diverse and extensive set of 4800 groups, combining various subjects (objects, plants, animals, human actions, textures) with multiple styles. SMA enables flexible control over groups and classes, allowing us to configure datasets to reflect diverse benchmark scenarios. While ideally, data collection would capture extensive group diversity, practical constraints often make this infeasible. SMA addresses this by enabling large and configurable group structures through flexible control over styles, subject classes, and domains-allowing datasets to reflect a wide range of real-world benchmark scenarios. This design not only expands group and class diversity, but also opens new methodological directions for evaluating model performance across diverse group and domain configurations-including scenarios with many minority groups, varying group imbalance, and complex domain shifts-and for studying fairness, robustness, and adaptation under a broader range of realistic conditions. To demonstrate SMA's effectiveness, we implemented two benchmarks: (1) a novel OOD generalization and group fairness benchmark leveraging SMA's domain, class, and group diversity to evaluate existing benchmarks. Our findings reveal that while simple balancing and algorithms utilizing group information remain competitive as claimed in previous benchmarks, increasing group diversity significantly impacts fairness, altering the superiority and relative rankings of algorithms. We also propose to use \textit{Top-M worst group accuracy} as a new hyperparameter tuning metric, demonstrating broader fairness during optimization and delivering better final worst-group accuracy for larger group diversity. (2) An unsupervised domain adaptation (UDA) benchmark utilizing SMA's group diversity to evaluate UDA algorithms across more scenarios, offering a more comprehensive benchmark with lower error bars (reduced by 73\% and 28\% in closed-set setting and UniDA setting, respectively) compared to existing efforts. These use cases highlight SMA's potential to significantly impact the outcomes of conventional benchmarks.
3DPyranet Features Fusion for Spatio-temporal Feature Learning
Apr 26, 2025Abstract:Convolutional neural network (CNN) slides a kernel over the whole image to produce an output map. This kernel scheme reduces the number of parameters with respect to a fully connected neural network (NN). While CNN has proven to be an effective model in recognition of handwritten characters and traffic signal sign boards, etc. recently, its deep variants have proven to be effective in similar as well as more challenging applications like object, scene and action recognition. Deep CNN add more layers and kernels to the classical CNN, increasing the number of parameters, and partly reducing the main advantage of CNN which is less parameters. In this paper, a 3D pyramidal neural network called 3DPyraNet and a discriminative approach for spatio-temporal feature learning based on it, called 3DPyraNet-F, are proposed. 3DPyraNet introduces a new weighting scheme which learns features from both spatial and temporal dimensions analyzing multiple adjacent frames and keeping a biological plausible structure. It keeps the spatial topology of the input image and presents fewer parameters and lower computational and memory costs compared to both fully connected NNs and recent deep CNNs. 3DPyraNet-F extract the features maps of the highest layer of the learned network, fuse them in a single vector, and provide it as input in such a way to a linear-SVM classifier that enhances the recognition of human actions and dynamic scenes from the videos. Encouraging results are reported with 3DPyraNet in real-world environments, especially in the presence of camera induced motion. Further, 3DPyraNet-F clearly outperforms the state-of-the-art on three benchmark datasets and shows comparable result for the fourth.
Point Tracking in Surgery--The 2024 Surgical Tattoos in Infrared (STIR) Challenge
Mar 31, 2025Abstract:Understanding tissue motion in surgery is crucial to enable applications in downstream tasks such as segmentation, 3D reconstruction, virtual tissue landmarking, autonomous probe-based scanning, and subtask autonomy. Labeled data are essential to enabling algorithms in these downstream tasks since they allow us to quantify and train algorithms. This paper introduces a point tracking challenge to address this, wherein participants can submit their algorithms for quantification. The submitted algorithms are evaluated using a dataset named surgical tattoos in infrared (STIR), with the challenge aptly named the STIR Challenge 2024. The STIR Challenge 2024 comprises two quantitative components: accuracy and efficiency. The accuracy component tests the accuracy of algorithms on in vivo and ex vivo sequences. The efficiency component tests the latency of algorithm inference. The challenge was conducted as a part of MICCAI EndoVis 2024. In this challenge, we had 8 total teams, with 4 teams submitting before and 4 submitting after challenge day. This paper details the STIR Challenge 2024, which serves to move the field towards more accurate and efficient algorithms for spatial understanding in surgery. In this paper we summarize the design, submissions, and results from the challenge. The challenge dataset is available here: https://zenodo.org/records/14803158 , and the code for baseline models and metric calculation is available here: https://github.com/athaddius/STIRMetrics
Video-DPRP: A Differentially Private Approach for Visual Privacy-Preserving Video Human Activity Recognition
Mar 03, 2025Abstract:Considerable effort has been made in privacy-preserving video human activity recognition (HAR). Two primary approaches to ensure privacy preservation in Video HAR are differential privacy (DP) and visual privacy. Techniques enforcing DP during training provide strong theoretical privacy guarantees but offer limited capabilities for visual privacy assessment. Conversely methods, such as low-resolution transformations, data obfuscation and adversarial networks, emphasize visual privacy but lack clear theoretical privacy assurances. In this work, we focus on two main objectives: (1) leveraging DP properties to develop a model-free approach for visual privacy in videos and (2) evaluating our proposed technique using both differential privacy and visual privacy assessments on HAR tasks. To achieve goal (1), we introduce Video-DPRP: a Video-sample-wise Differentially Private Random Projection framework for privacy-preserved video reconstruction for HAR. By using random projections, noise matrices and right singular vectors derived from the singular value decomposition of videos, Video-DPRP reconstructs DP videos using privacy parameters ($\epsilon,\delta$) while enabling visual privacy assessment. For goal (2), using UCF101 and HMDB51 datasets, we compare Video-DPRP's performance on activity recognition with traditional DP methods, and state-of-the-art (SOTA) visual privacy-preserving techniques. Additionally, we assess its effectiveness in preserving privacy-related attributes such as facial features, gender, and skin color, using the PA-HMDB and VISPR datasets. Video-DPRP combines privacy-preservation from both a DP and visual privacy perspective unlike SOTA methods that typically address only one of these aspects.
Usefulness of LLMs as an Author Checklist Assistant for Scientific Papers: NeurIPS'24 Experiment
Nov 05, 2024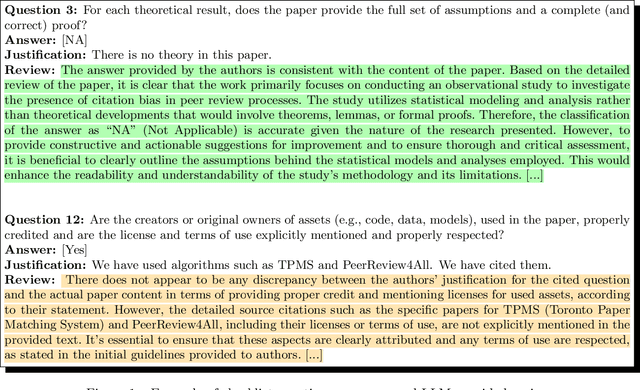
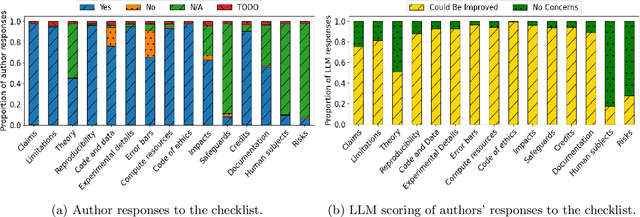
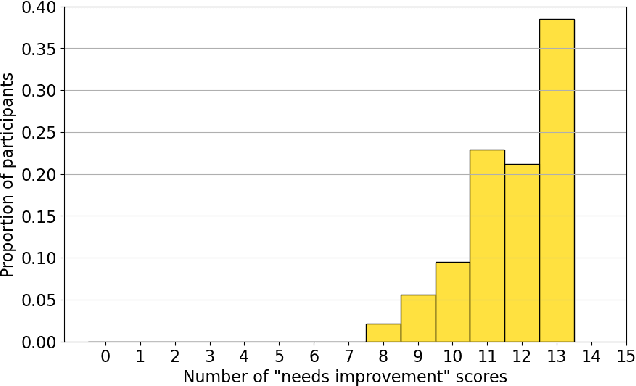

Abstract:Large language models (LLMs) represent a promising, but controversial, tool in aiding scientific peer review. This study evaluates the usefulness of LLMs in a conference setting as a tool for vetting paper submissions against submission standards. We conduct an experiment at the 2024 Neural Information Processing Systems (NeurIPS) conference, where 234 papers were voluntarily submitted to an "LLM-based Checklist Assistant." This assistant validates whether papers adhere to the author checklist used by NeurIPS, which includes questions to ensure compliance with research and manuscript preparation standards. Evaluation of the assistant by NeurIPS paper authors suggests that the LLM-based assistant was generally helpful in verifying checklist completion. In post-usage surveys, over 70% of authors found the assistant useful, and 70% indicate that they would revise their papers or checklist responses based on its feedback. While causal attribution to the assistant is not definitive, qualitative evidence suggests that the LLM contributed to improving some submissions. Survey responses and analysis of re-submissions indicate that authors made substantive revisions to their submissions in response to specific feedback from the LLM. The experiment also highlights common issues with LLMs: inaccuracy (20/52) and excessive strictness (14/52) were the most frequent issues flagged by authors. We also conduct experiments to understand potential gaming of the system, which reveal that the assistant could be manipulated to enhance scores through fabricated justifications, highlighting potential vulnerabilities of automated review tools.
FAIR Universe HiggsML Uncertainty Challenge Competition
Oct 03, 2024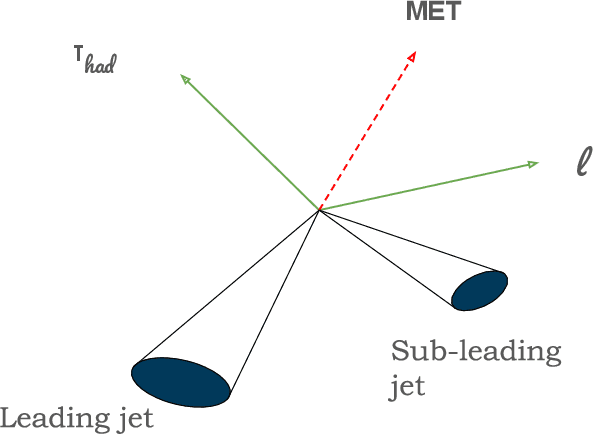
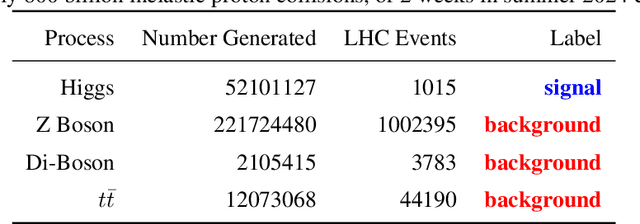
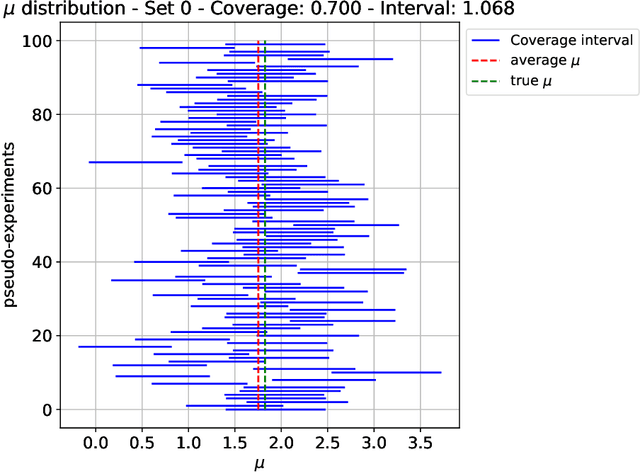
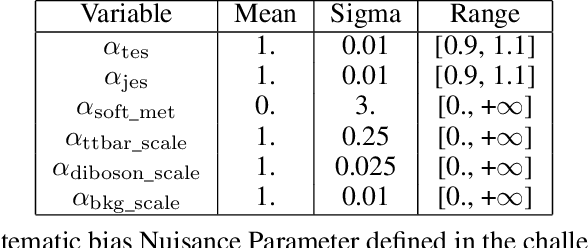
Abstract:The FAIR Universe -- HiggsML Uncertainty Challenge focuses on measuring the physics properties of elementary particles with imperfect simulators due to differences in modelling systematic errors. Additionally, the challenge is leveraging a large-compute-scale AI platform for sharing datasets, training models, and hosting machine learning competitions. Our challenge brings together the physics and machine learning communities to advance our understanding and methodologies in handling systematic (epistemic) uncertainties within AI techniques.
RelevAI-Reviewer: A Benchmark on AI Reviewers for Survey Paper Relevance
Jun 13, 2024



Abstract:Recent advancements in Artificial Intelligence (AI), particularly the widespread adoption of Large Language Models (LLMs), have significantly enhanced text analysis capabilities. This technological evolution offers considerable promise for automating the review of scientific papers, a task traditionally managed through peer review by fellow researchers. Despite its critical role in maintaining research quality, the conventional peer-review process is often slow and subject to biases, potentially impeding the swift propagation of scientific knowledge. In this paper, we propose RelevAI-Reviewer, an automatic system that conceptualizes the task of survey paper review as a classification problem, aimed at assessing the relevance of a paper in relation to a specified prompt, analogous to a "call for papers". To address this, we introduce a novel dataset comprised of 25,164 instances. Each instance contains one prompt and four candidate papers, each varying in relevance to the prompt. The objective is to develop a machine learning (ML) model capable of determining the relevance of each paper and identifying the most pertinent one. We explore various baseline approaches, including traditional ML classifiers like Support Vector Machine (SVM) and advanced language models such as BERT. Preliminary findings indicate that the BERT-based end-to-end classifier surpasses other conventional ML methods in performance. We present this problem as a public challenge to foster engagement and interest in this area of research.
Depth Estimation using Weighted-loss and Transfer Learning
Apr 11, 2024



Abstract:Depth estimation from 2D images is a common computer vision task that has applications in many fields including autonomous vehicles, scene understanding and robotics. The accuracy of a supervised depth estimation method mainly relies on the chosen loss function, the model architecture, quality of data and performance metrics. In this study, we propose a simplified and adaptable approach to improve depth estimation accuracy using transfer learning and an optimized loss function. The optimized loss function is a combination of weighted losses to which enhance robustness and generalization: Mean Absolute Error (MAE), Edge Loss and Structural Similarity Index (SSIM). We use a grid search and a random search method to find optimized weights for the losses, which leads to an improved model. We explore multiple encoder-decoder-based models including DenseNet121, DenseNet169, DenseNet201, and EfficientNet for the supervised depth estimation model on NYU Depth Dataset v2. We observe that the EfficientNet model, pre-trained on ImageNet for classification when used as an encoder, with a simple upsampling decoder, gives the best results in terms of RSME, REL and log10: 0.386, 0.113 and 0.049, respectively. We also perform a qualitative analysis which illustrates that our model produces depth maps that closely resemble ground truth, even in cases where the ground truth is flawed. The results indicate significant improvements in accuracy and robustness, with EfficientNet being the most successful architecture.
Beyond the Known: Adversarial Autoencoders in Novelty Detection
Apr 06, 2024Abstract:In novelty detection, the goal is to decide if a new data point should be categorized as an inlier or an outlier, given a training dataset that primarily captures the inlier distribution. Recent approaches typically use deep encoder and decoder network frameworks to derive a reconstruction error, and employ this error either to determine a novelty score, or as the basis for a one-class classifier. In this research, we use a similar framework but with a lightweight deep network, and we adopt a probabilistic score with reconstruction error. Our methodology calculates the probability of whether the sample comes from the inlier distribution or not. This work makes two key contributions. The first is that we compute the novelty probability by linearizing the manifold that holds the structure of the inlier distribution. This allows us to interpret how the probability is distributed and can be determined in relation to the local coordinates of the manifold tangent space. The second contribution is that we improve the training protocol for the network. Our results indicate that our approach is effective at learning the target class, and it outperforms recent state-of-the-art methods on several benchmark datasets.
Synthesizing CTA Image Data for Type-B Aortic Dissection using Stable Diffusion Models
Feb 10, 2024Abstract:Stable Diffusion (SD) has gained a lot of attention in recent years in the field of Generative AI thus helping in synthesizing medical imaging data with distinct features. The aim is to contribute to the ongoing effort focused on overcoming the limitations of data scarcity and improving the capabilities of ML algorithms for cardiovascular image processing. Therefore, in this study, the possibility of generating synthetic cardiac CTA images was explored by fine-tuning stable diffusion models based on user defined text prompts, using only limited number of CTA images as input. A comprehensive evaluation of the synthetic data was conducted by incorporating both quantitative analysis and qualitative assessment, where a clinician assessed the quality of the generated data. It has been shown that Cardiac CTA images can be successfully generated using using Text to Image (T2I) stable diffusion model. The results demonstrate that the tuned T2I CTA diffusion model was able to generate images with features that are typically unique to acute type B aortic dissection (TBAD) medical conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge