Mei Liu
N3C Natural Language Processing
User-Adaptive Meta-Learning for Cold-Start Medication Recommendation with Uncertainty Filtering
Jan 30, 2026Abstract:Large-scale Electronic Health Record (EHR) databases have become indispensable in supporting clinical decision-making through data-driven treatment recommendations. However, existing medication recommender methods often struggle with a user (i.e., patient) cold-start problem, where recommendations for new patients are usually unreliable due to the lack of sufficient prescription history for patient profiling. While prior studies have utilized medical knowledge graphs to connect medication concepts through pharmacological or chemical relationships, these methods primarily focus on mitigating the item cold-start issue and fall short in providing personalized recommendations that adapt to individual patient characteristics. Meta-learning has shown promise in handling new users with sparse interactions in recommender systems. However, its application to EHRs remains underexplored due to the unique sequential structure of EHR data. To tackle these challenges, we propose MetaDrug, a multi-level, uncertainty-aware meta-learning framework designed to address the patient cold-start problem in medication recommendation. MetaDrug proposes a novel two-level meta-adaptation mechanism, including self-adaptation, which adapts the model to new patients using their own medical events as support sets to capture temporal dependencies; and peer-adaptation, which adapts the model using similar visits from peer patients to enrich new patient representations. Meanwhile, to further improve meta-adaptation outcomes, we introduce an uncertainty quantification module that ranks the support visits and filters out the unrelated information for adaptation consistency. We evaluate our approach on the MIMIC-III and Acute Kidney Injury (AKI) datasets. Experimental results on both datasets demonstrate that MetaDrug consistently outperforms state-of-the-art medication recommendation methods on cold-start patients.
SurvAttack: Black-Box Attack On Survival Models through Ontology-Informed EHR Perturbation
Dec 24, 2024



Abstract:Survival analysis (SA) models have been widely studied in mining electronic health records (EHRs), particularly in forecasting the risk of critical conditions for prioritizing high-risk patients. However, their vulnerability to adversarial attacks is much less explored in the literature. Developing black-box perturbation algorithms and evaluating their impact on state-of-the-art survival models brings two benefits to medical applications. First, it can effectively evaluate the robustness of models in pre-deployment testing. Also, exploring how subtle perturbations would result in significantly different outcomes can provide counterfactual insights into the clinical interpretation of model prediction. In this work, we introduce SurvAttack, a novel black-box adversarial attack framework leveraging subtle clinically compatible, and semantically consistent perturbations on longitudinal EHRs to degrade survival models' predictive performance. We specifically develop a greedy algorithm to manipulate medical codes with various adversarial actions throughout a patient's medical history. Then, these adversarial actions are prioritized using a composite scoring strategy based on multi-aspect perturbation quality, including saliency, perturbation stealthiness, and clinical meaningfulness. The proposed adversarial EHR perturbation algorithm is then used in an efficient SA-specific strategy to attack a survival model when estimating the temporal ranking of survival urgency for patients. To demonstrate the significance of our work, we conduct extensive experiments, including baseline comparisons, explainability analysis, and case studies. The experimental results affirm our research's effectiveness in illustrating the vulnerabilities of patient survival models, model interpretation, and ultimately contributing to healthcare quality.
First-frame Supervised Video Polyp Segmentation via Propagative and Semantic Dual-teacher Network
Dec 21, 2024Abstract:Automatic video polyp segmentation plays a critical role in gastrointestinal cancer screening, but the cost of frameby-frame annotations is prohibitively high. While sparse-frame supervised methods have reduced this burden proportionately, the cost remains overwhelming for long-duration videos and large-scale datasets. In this paper, we, for the first time, reduce the annotation cost to just a single frame per polyp video, regardless of the video's length. To this end, we introduce a new task, First-Frame Supervised Video Polyp Segmentation (FSVPS), and propose a novel Propagative and Semantic Dual-Teacher Network (PSDNet). Specifically, PSDNet adopts a teacher-student framework but employs two distinct types of teachers: the propagative teacher and the semantic teacher. The propagative teacher is a universal object tracker that propagates the first-frame annotation to subsequent frames as pseudo labels. However, tracking errors may accumulate over time, gradually degrading the pseudo labels and misguiding the student model. To address this, we introduce the semantic teacher, an exponential moving average of the student model, which produces more stable and time-invariant pseudo labels. PSDNet merges the pseudo labels from both teachers using a carefully-designed back-propagation strategy. This strategy assesses the quality of the pseudo labels by tracking them backward to the first frame. High-quality pseudo labels are more likely to spatially align with the firstframe annotation after this backward tracking, ensuring more accurate teacher-to-student knowledge transfer and improved segmentation performance. Benchmarking on SUN-SEG, the largest VPS dataset, demonstrates the competitive performance of PSDNet compared to fully-supervised approaches, and its superiority over sparse-frame supervised state-of-the-arts with a minimum improvement of 4.5% in Dice score.
Contrastive Learning on Medical Intents for Sequential Prescription Recommendation
Aug 13, 2024



Abstract:Recent advancements in sequential modeling applied to Electronic Health Records (EHR) have greatly influenced prescription recommender systems. While the recent literature on drug recommendation has shown promising performance, the study of discovering a diversity of coexisting temporal relationships at the level of medical codes over consecutive visits remains less explored. The goal of this study can be motivated from two perspectives. First, there is a need to develop a sophisticated sequential model capable of disentangling the complex relationships across sequential visits. Second, it is crucial to establish multiple and diverse health profiles for the same patient to ensure a comprehensive consideration of different medical intents in drug recommendation. To achieve this goal, we introduce Attentive Recommendation with Contrasted Intents (ARCI), a multi-level transformer-based method designed to capture the different but coexisting temporal paths across a shared sequence of visits. Specifically, we propose a novel intent-aware method with contrastive learning, that links specialized medical intents of the patients to the transformer heads for extracting distinct temporal paths associated with different health profiles. We conducted experiments on two real-world datasets for the prescription recommendation task using both ranking and classification metrics. Our results demonstrate that ARCI has outperformed the state-of-the-art prescription recommendation methods and is capable of providing interpretable insights for healthcare practitioners.
SALI: Short-term Alignment and Long-term Interaction Network for Colonoscopy Video Polyp Segmentation
Jun 19, 2024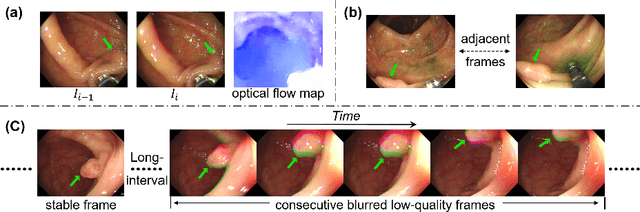
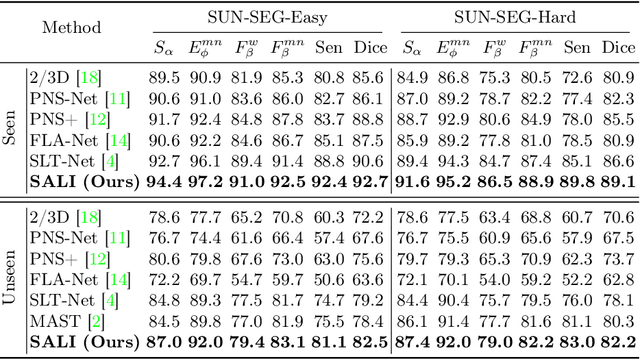
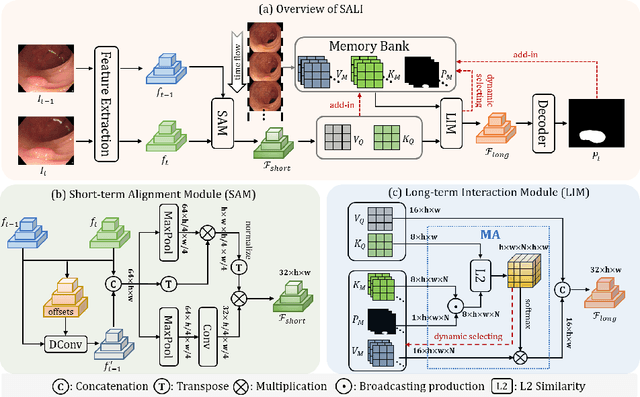
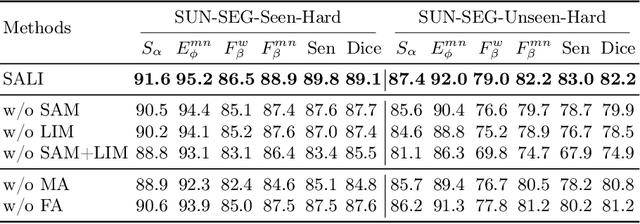
Abstract:Colonoscopy videos provide richer information in polyp segmentation for rectal cancer diagnosis. However, the endoscope's fast moving and close-up observing make the current methods suffer from large spatial incoherence and continuous low-quality frames, and thus yield limited segmentation accuracy. In this context, we focus on robust video polyp segmentation by enhancing the adjacent feature consistency and rebuilding the reliable polyp representation. To achieve this goal, we in this paper propose SALI network, a hybrid of Short-term Alignment Module (SAM) and Long-term Interaction Module (LIM). The SAM learns spatial-aligned features of adjacent frames via deformable convolution and further harmonizes them to capture more stable short-term polyp representation. In case of low-quality frames, the LIM stores the historical polyp representations as a long-term memory bank, and explores the retrospective relations to interactively rebuild more reliable polyp features for the current segmentation. Combing SAM and LIM, the SALI network of video segmentation shows a great robustness to the spatial variations and low-visual cues. Benchmark on the large-scale SUNSEG verifies the superiority of SALI over the current state-of-the-arts by improving Dice by 2.1%, 2.5%, 4.1% and 1.9%, for the four test sub-sets, respectively. Codes are at https://github.com/Scatteredrain/SALI.
MonoPCC: Photometric-invariant Cycle Constraint for Monocular Depth Estimation of Endoscopic Images
Apr 25, 2024
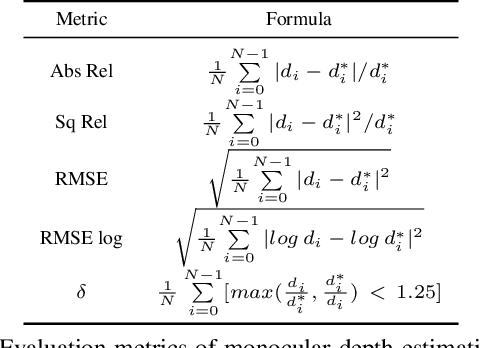
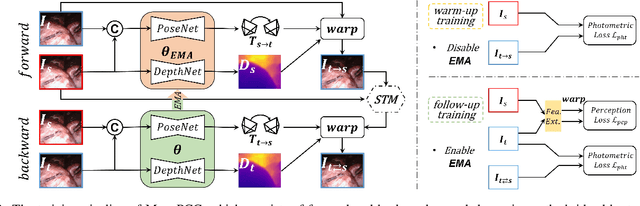
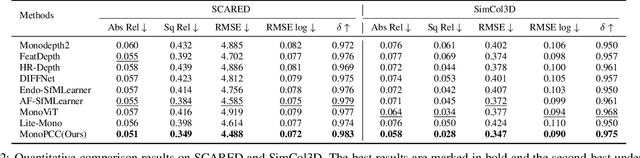
Abstract:Photometric constraint is indispensable for self-supervised monocular depth estimation. It involves warping a source image onto a target view using estimated depth&pose, and then minimizing the difference between the warped and target images. However, the endoscopic built-in light causes significant brightness fluctuations, and thus makes the photometric constraint unreliable. Previous efforts only mitigate this relying on extra models to calibrate image brightness. In this paper, we propose MonoPCC to address the brightness inconsistency radically by reshaping the photometric constraint into a cycle form. Instead of only warping the source image, MonoPCC constructs a closed loop consisting of two opposite forward-backward warping paths: from target to source and then back to target. Thus, the target image finally receives an image cycle-warped from itself, which naturally makes the constraint invariant to brightness changes. Moreover, MonoPCC transplants the source image's phase-frequency into the intermediate warped image to avoid structure lost, and also stabilizes the training via an exponential moving average (EMA) strategy to avoid frequent changes in the forward warping. The comprehensive and extensive experimental results on three datasets demonstrate that our proposed MonoPCC shows a great robustness to the brightness inconsistency, and exceeds other state-of-the-arts by reducing the absolute relative error by at least 7.27%.
MonoBox: Tightness-free Box-supervised Polyp Segmentation using Monotonicity Constraint
Apr 02, 2024



Abstract:We propose MonoBox, an innovative box-supervised segmentation method constrained by monotonicity to liberate its training from the user-unfriendly box-tightness assumption. In contrast to conventional box-supervised segmentation, where the box edges must precisely touch the target boundaries, MonoBox leverages imprecisely-annotated boxes to achieve robust pixel-wise segmentation. The 'linchpin' is that, within the noisy zones around box edges, MonoBox discards the traditional misguiding multiple-instance learning loss, and instead optimizes a carefully-designed objective, termed monotonicity constraint. Along directions transitioning from the foreground to background, this new constraint steers responses to adhere to a trend of monotonically decreasing values. Consequently, the originally unreliable learning within the noisy zones is transformed into a correct and effective monotonicity optimization. Moreover, an adaptive label correction is introduced, enabling MonoBox to enhance the tightness of box annotations using predicted masks from the previous epoch and dynamically shrink the noisy zones as training progresses. We verify MonoBox in the box-supervised segmentation task of polyps, where satisfying box-tightness is challenging due to the vague boundaries between the polyp and normal tissues. Experiments on both public synthetic and in-house real noisy datasets demonstrate that MonoBox exceeds other anti-noise state-of-the-arts by improving Dice by at least 5.5% and 3.3%, respectively. Codes are at https://github.com/Huster-Hq/MonoBox.
IoT Network Traffic Analysis with Deep Learning
Feb 06, 2024



Abstract:As IoT networks become more complex and generate massive amounts of dynamic data, it is difficult to monitor and detect anomalies using traditional statistical methods and machine learning methods. Deep learning algorithms can process and learn from large amounts of data and can also be trained using unsupervised learning techniques, meaning they don't require labelled data to detect anomalies. This makes it possible to detect new and unknown anomalies that may not have been detected before. Also, deep learning algorithms can be automated and highly scalable; thereby, they can run continuously in the backend and make it achievable to monitor large IoT networks instantly. In this work, we conduct a literature review on the most recent works using deep learning techniques and implement a model using ensemble techniques on the KDD Cup 99 dataset. The experimental results showcase the impressive performance of our deep anomaly detection model, achieving an accuracy of over 98\%.
IBoxCLA: Towards Robust Box-supervised Segmentation of Polyp via Improved Box-dice and Contrastive Latent-anchors
Oct 14, 2023



Abstract:Box-supervised polyp segmentation attracts increasing attention for its cost-effective potential. Existing solutions often rely on learning-free methods or pretrained models to laboriously generate pseudo masks, triggering Dice constraint subsequently. In this paper, we found that a model guided by the simplest box-filled masks can accurately predict polyp locations/sizes, but suffers from shape collapsing. In response, we propose two innovative learning fashions, Improved Box-dice (IBox) and Contrastive Latent-Anchors (CLA), and combine them to train a robust box-supervised model IBoxCLA. The core idea behind IBoxCLA is to decouple the learning of location/size and shape, allowing for focused constraints on each of them. Specifically, IBox transforms the segmentation map into a proxy map using shape decoupling and confusion-region swapping sequentially. Within the proxy map, shapes are disentangled, while locations/sizes are encoded as box-like responses. By constraining the proxy map instead of the raw prediction, the box-filled mask can well supervise IBoxCLA without misleading its shape learning. Furthermore, CLA contributes to shape learning by generating two types of latent anchors, which are learned and updated using momentum and segmented polyps to steadily represent polyp and background features. The latent anchors facilitate IBoxCLA to capture discriminative features within and outside boxes in a contrastive manner, yielding clearer boundaries. We benchmark IBoxCLA on five public polyp datasets. The experimental results demonstrate the competitive performance of IBoxCLA compared to recent fully-supervised polyp segmentation methods, and its superiority over other box-supervised state-of-the-arts with a relative increase of overall mDice and mIoU by at least 6.5% and 7.5%, respectively.
Contrastive Learning of Temporal Distinctiveness for Survival Analysis in Electronic Health Records
Aug 24, 2023



Abstract:Survival analysis plays a crucial role in many healthcare decisions, where the risk prediction for the events of interest can support an informative outlook for a patient's medical journey. Given the existence of data censoring, an effective way of survival analysis is to enforce the pairwise temporal concordance between censored and observed data, aiming to utilize the time interval before censoring as partially observed time-to-event labels for supervised learning. Although existing studies mostly employed ranking methods to pursue an ordering objective, contrastive methods which learn a discriminative embedding by having data contrast against each other, have not been explored thoroughly for survival analysis. Therefore, in this paper, we propose a novel Ontology-aware Temporality-based Contrastive Survival (OTCSurv) analysis framework that utilizes survival durations from both censored and observed data to define temporal distinctiveness and construct negative sample pairs with adjustable hardness for contrastive learning. Specifically, we first use an ontological encoder and a sequential self-attention encoder to represent the longitudinal EHR data with rich contexts. Second, we design a temporal contrastive loss to capture varying survival durations in a supervised setting through a hardness-aware negative sampling mechanism. Last, we incorporate the contrastive task into the time-to-event predictive task with multiple loss components. We conduct extensive experiments using a large EHR dataset to forecast the risk of hospitalized patients who are in danger of developing acute kidney injury (AKI), a critical and urgent medical condition. The effectiveness and explainability of the proposed model are validated through comprehensive quantitative and qualitative studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge