Ramakanth Kavuluru
N3C Natural Language Processing
Large Language Models for Drug Overdose Prediction from Longitudinal Medical Records
Apr 16, 2025Abstract:The ability to predict drug overdose risk from a patient's medical records is crucial for timely intervention and prevention. Traditional machine learning models have shown promise in analyzing longitudinal medical records for this task. However, recent advancements in large language models (LLMs) offer an opportunity to enhance prediction performance by leveraging their ability to process long textual data and their inherent prior knowledge across diverse tasks. In this study, we assess the effectiveness of Open AI's GPT-4o LLM in predicting drug overdose events using patients' longitudinal insurance claims records. We evaluate its performance in both fine-tuned and zero-shot settings, comparing them to strong traditional machine learning methods as baselines. Our results show that LLMs not only outperform traditional models in certain settings but can also predict overdose risk in a zero-shot setting without task-specific training. These findings highlight the potential of LLMs in clinical decision support, particularly for drug overdose risk prediction.
A Benchmark for End-to-End Zero-Shot Biomedical Relation Extraction with LLMs: Experiments with OpenAI Models
Apr 05, 2025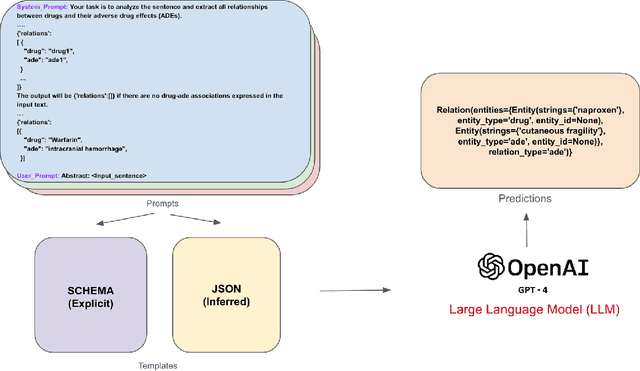
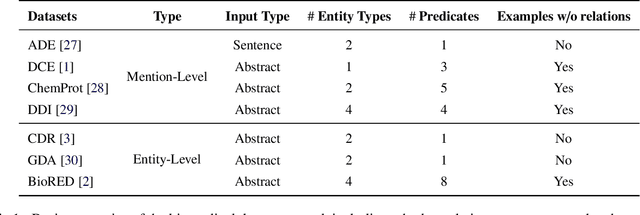
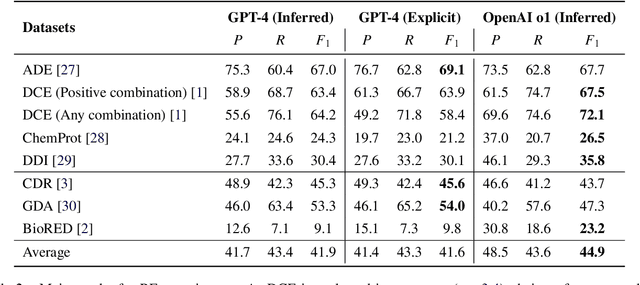
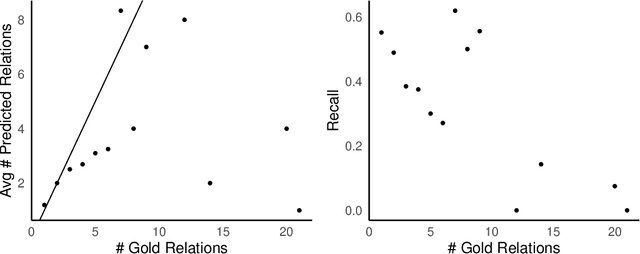
Abstract:Objective: Zero-shot methodology promises to cut down on costs of dataset annotation and domain expertise needed to make use of NLP. Generative large language models trained to align with human goals have achieved high zero-shot performance across a wide variety of tasks. As of yet, it is unclear how well these models perform on biomedical relation extraction (RE). To address this knowledge gap, we explore patterns in the performance of OpenAI LLMs across a diverse sampling of RE tasks. Methods: We use OpenAI GPT-4-turbo and their reasoning model o1 to conduct end-to-end RE experiments on seven datasets. We use the JSON generation capabilities of GPT models to generate structured output in two ways: (1) by defining an explicit schema describing the structure of relations, and (2) using a setting that infers the structure from the prompt language. Results: Our work is the first to study and compare the performance of the GPT-4 and o1 for the end-to-end zero-shot biomedical RE task across a broad array of datasets. We found the zero-shot performances to be proximal to that of fine-tuned methods. The limitations of this approach are that it performs poorly on instances containing many relations and errs on the boundaries of textual mentions. Conclusion: Recent large language models exhibit promising zero-shot capabilities in complex biomedical RE tasks, offering competitive performance with reduced dataset curation and NLP modeling needs at the cost of increased computing, potentially increasing medical community accessibility. Addressing the limitations we identify could further boost reliability. The code, data, and prompts for all our experiments are publicly available: https://github.com/bionlproc/ZeroShotRE
Do LLMs Surpass Encoders for Biomedical NER?
Apr 01, 2025Abstract:Recognizing spans of biomedical concepts and their types (e.g., drug or gene) in free text, often called biomedical named entity recognition (NER), is a basic component of information extraction (IE) pipelines. Without a strong NER component, other applications, such as knowledge discovery and information retrieval, are not practical. State-of-the-art in NER shifted from traditional ML models to deep neural networks with transformer-based encoder models (e.g., BERT) emerging as the current standard. However, decoder models (also called large language models or LLMs) are gaining traction in IE. But LLM-driven NER often ignores positional information due to the generative nature of decoder models. Furthermore, they are computationally very expensive (both in inference time and hardware needs). Hence, it is worth exploring if they actually excel at biomedical NER and assess any associated trade-offs (performance vs efficiency). This is exactly what we do in this effort employing the same BIO entity tagging scheme (that retains positional information) using five different datasets with varying proportions of longer entities. Our results show that the LLMs chosen (Mistral and Llama: 8B range) often outperform best encoder models (BERT-(un)cased, BiomedBERT, and DeBERTav3: 300M range) by 2-8% in F-scores except for one dataset, where they equal encoder performance. This gain is more prominent among longer entities of length >= 3 tokens. However, LLMs are one to two orders of magnitude more expensive at inference time and may need cost prohibitive hardware. Thus, when performance differences are small or real time user feedback is needed, encoder models might still be more suitable than LLMs.
Clinical Reading Comprehension with Encoder-Decoder Models Enhanced by Direct Preference Optimization
Jul 19, 2024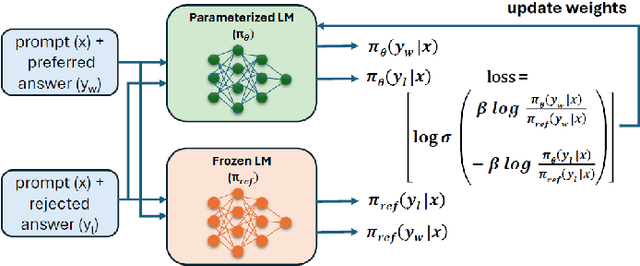
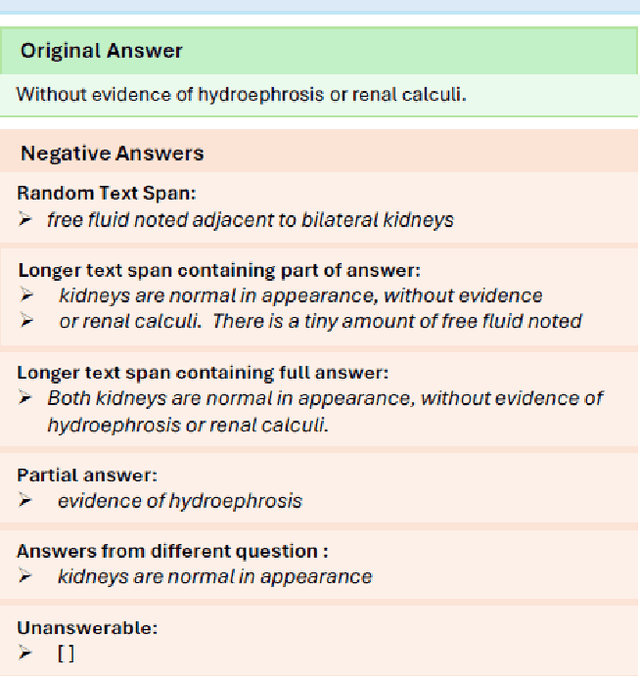
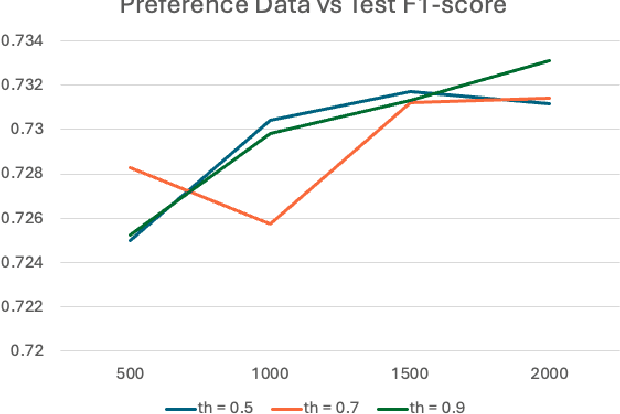
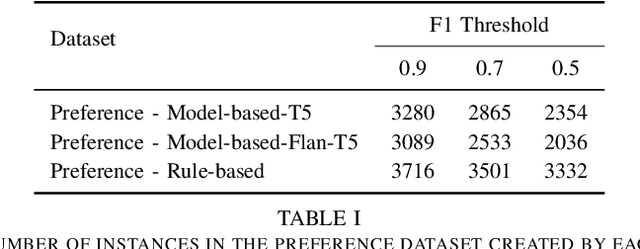
Abstract:Extractive question answering over clinical text is a crucial need to help deal with the deluge of clinical text generated in hospitals. While encoder models (e.g., BERT) have been popular for this reading comprehension task, recently encoder-decoder models (e.g., T5) are on the rise. There is also the emergence of preference optimization techniques to align decoder-only LLMs with human preferences. In this paper, we combine encoder-decoder models with the direct preference optimization (DPO) method to improve over prior state of the art for the RadQA radiology question answering task by 12-15 F1 points. To the best of our knowledge, this effort is the first to show that DPO method also works for reading comprehension via novel heuristics to generate preference data without human inputs.
Knowledge-Driven Cross-Document Relation Extraction
May 22, 2024Abstract:Relation extraction (RE) is a well-known NLP application often treated as a sentence- or document-level task. However, a handful of recent efforts explore it across documents or in the cross-document setting (CrossDocRE). This is distinct from the single document case because different documents often focus on disparate themes, while text within a document tends to have a single goal. Linking findings from disparate documents to identify new relationships is at the core of the popular literature-based knowledge discovery paradigm in biomedicine and other domains. Current CrossDocRE efforts do not consider domain knowledge, which are often assumed to be known to the reader when documents are authored. Here, we propose a novel approach, KXDocRE, that embed domain knowledge of entities with input text for cross-document RE. Our proposed framework has three main benefits over baselines: 1) it incorporates domain knowledge of entities along with documents' text; 2) it offers interpretability by producing explanatory text for predicted relations between entities 3) it improves performance over the prior methods.
How Important is Domain Specificity in Language Models and Instruction Finetuning for Biomedical Relation Extraction?
Feb 21, 2024Abstract:Cutting edge techniques developed in the general NLP domain are often subsequently applied to the high-value, data-rich biomedical domain. The past few years have seen generative language models (LMs), instruction finetuning, and few-shot learning become foci of NLP research. As such, generative LMs pretrained on biomedical corpora have proliferated and biomedical instruction finetuning has been attempted as well, all with the hope that domain specificity improves performance on downstream tasks. Given the nontrivial effort in training such models, we investigate what, if any, benefits they have in the key biomedical NLP task of relation extraction. Specifically, we address two questions: (1) Do LMs trained on biomedical corpora outperform those trained on general domain corpora? (2) Do models instruction finetuned on biomedical datasets outperform those finetuned on assorted datasets or those simply pretrained? We tackle these questions using existing LMs, testing across four datasets. In a surprising result, general-domain models typically outperformed biomedical-domain models. However, biomedical instruction finetuning improved performance to a similar degree as general instruction finetuning, despite having orders of magnitude fewer instructions. Our findings suggest it may be more fruitful to focus research effort on larger-scale biomedical instruction finetuning of general LMs over building domain-specific biomedical LMs
Revisiting Document-Level Relation Extraction with Context-Guided Link Prediction
Jan 22, 2024Abstract:Document-level relation extraction (DocRE) poses the challenge of identifying relationships between entities within a document as opposed to the traditional RE setting where a single sentence is input. Existing approaches rely on logical reasoning or contextual cues from entities. This paper reframes document-level RE as link prediction over a knowledge graph with distinct benefits: 1) Our approach combines entity context with document-derived logical reasoning, enhancing link prediction quality. 2) Predicted links between entities offer interpretability, elucidating employed reasoning. We evaluate our approach on three benchmark datasets: DocRED, ReDocRED, and DWIE. The results indicate that our proposed method outperforms the state-of-the-art models and suggests that incorporating context-based link prediction techniques can enhance the performance of document-level relation extraction models.
Comparison of pipeline, sequence-to-sequence, and GPT models for end-to-end relation extraction: experiments with the rare disease use-case
Nov 22, 2023Abstract:End-to-end relation extraction (E2ERE) is an important and realistic application of natural language processing (NLP) in biomedicine. In this paper, we aim to compare three prevailing paradigms for E2ERE using a complex dataset focused on rare diseases involving discontinuous and nested entities. We use the RareDis information extraction dataset to evaluate three competing approaches (for E2ERE): NER $\rightarrow$ RE pipelines, joint sequence to sequence models, and generative pre-trained transformer (GPT) models. We use comparable state-of-the-art models and best practices for each of these approaches and conduct error analyses to assess their failure modes. Our findings reveal that pipeline models are still the best, while sequence-to-sequence models are not far behind; GPT models with eight times as many parameters are worse than even sequence-to-sequence models and lose to pipeline models by over 10 F1 points. Partial matches and discontinuous entities caused many NER errors contributing to lower overall E2E performances. We also verify these findings on a second E2ERE dataset for chemical-protein interactions. Although generative LM-based methods are more suitable for zero-shot settings, when training data is available, our results show that it is better to work with more conventional models trained and tailored for E2ERE. More innovative methods are needed to marry the best of the both worlds from smaller encoder-decoder pipeline models and the larger GPT models to improve E2ERE. As of now, we see that well designed pipeline models offer substantial performance gains at a lower cost and carbon footprint for E2ERE. Our contribution is also the first to conduct E2ERE for the RareDis dataset.
End-to-End Models for Chemical-Protein Interaction Extraction: Better Tokenization and Span-Based Pipeline Strategies
Apr 03, 2023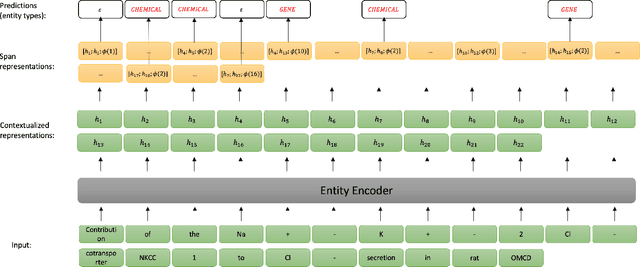
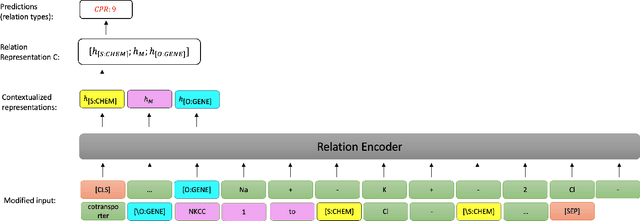
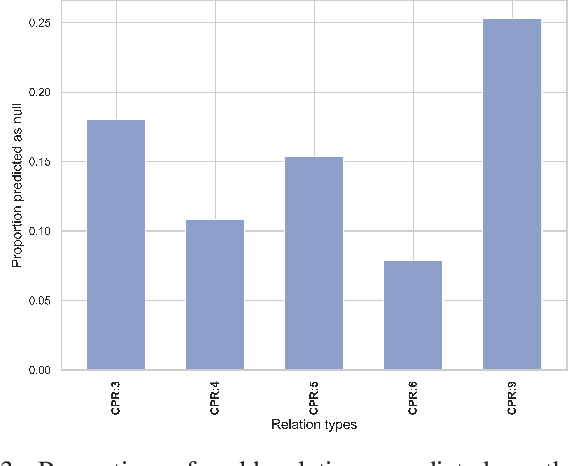
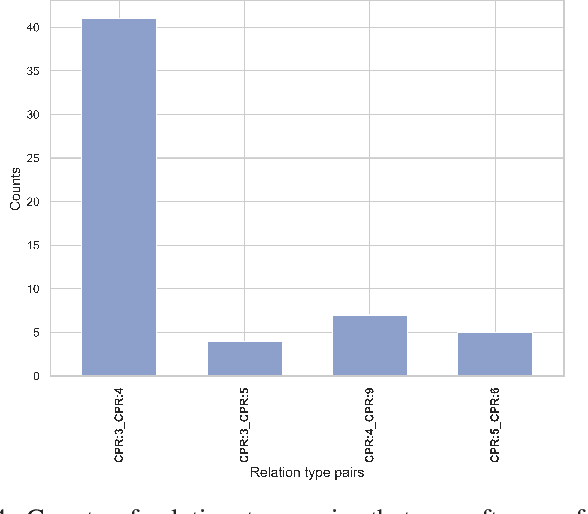
Abstract:End-to-end relation extraction (E2ERE) is an important task in information extraction, more so for biomedicine as scientific literature continues to grow exponentially. E2ERE typically involves identifying entities (or named entity recognition (NER)) and associated relations, while most RE tasks simply assume that the entities are provided upfront and end up performing relation classification. E2ERE is inherently more difficult than RE alone given the potential snowball effect of errors from NER leading to more errors in RE. A complex dataset in biomedical E2ERE is the ChemProt dataset (BioCreative VI, 2017) that identifies relations between chemical compounds and genes/proteins in scientific literature. ChemProt is included in all recent biomedical natural language processing benchmarks including BLUE, BLURB, and BigBio. However, its treatment in these benchmarks and in other separate efforts is typically not end-to-end, with few exceptions. In this effort, we employ a span-based pipeline approach to produce a new state-of-the-art E2ERE performance on the ChemProt dataset, resulting in $> 4\%$ improvement in F1-score over the prior best effort. Our results indicate that a straightforward fine-grained tokenization scheme helps span-based approaches excel in E2ERE, especially with regards to handling complex named entities. Our error analysis also identifies a few key failure modes in E2ERE for ChemProt.
End-to-End $n$-ary Relation Extraction for Combination Drug Therapies
Mar 29, 2023



Abstract:Combination drug therapies are treatment regimens that involve two or more drugs, administered more commonly for patients with cancer, HIV, malaria, or tuberculosis. Currently there are over 350K articles in PubMed that use the "combination drug therapy" MeSH heading with at least 10K articles published per year over the past two decades. Extracting combination therapies from scientific literature inherently constitutes an $n$-ary relation extraction problem. Unlike in the general $n$-ary setting where $n$ is fixed (e.g., drug-gene-mutation relations where $n=3$), extracting combination therapies is a special setting where $n \geq 2$ is dynamic, depending on each instance. Recently, Tiktinsky et al. (NAACL 2022) introduced a first of its kind dataset, CombDrugExt, for extracting such therapies from literature. Here, we use a sequence-to-sequence style end-to-end extraction method to achieve an F1-Score of $66.7\%$ on the CombDrugExt test set for positive (or effective) combinations. This is an absolute $\approx 5\%$ F1-score improvement even over the prior best relation classification score with spotted drug entities (hence, not end-to-end). Thus our effort introduces a state-of-the-art first model for end-to-end extraction that is already superior to the best prior non end-to-end model for this task. Our model seamlessly extracts all drug entities and relations in a single pass and is highly suitable for dynamic $n$-ary extraction scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge