Monika Jain
Knowledge-Driven Cross-Document Relation Extraction
May 22, 2024Abstract:Relation extraction (RE) is a well-known NLP application often treated as a sentence- or document-level task. However, a handful of recent efforts explore it across documents or in the cross-document setting (CrossDocRE). This is distinct from the single document case because different documents often focus on disparate themes, while text within a document tends to have a single goal. Linking findings from disparate documents to identify new relationships is at the core of the popular literature-based knowledge discovery paradigm in biomedicine and other domains. Current CrossDocRE efforts do not consider domain knowledge, which are often assumed to be known to the reader when documents are authored. Here, we propose a novel approach, KXDocRE, that embed domain knowledge of entities with input text for cross-document RE. Our proposed framework has three main benefits over baselines: 1) it incorporates domain knowledge of entities along with documents' text; 2) it offers interpretability by producing explanatory text for predicted relations between entities 3) it improves performance over the prior methods.
Revisiting Document-Level Relation Extraction with Context-Guided Link Prediction
Jan 22, 2024Abstract:Document-level relation extraction (DocRE) poses the challenge of identifying relationships between entities within a document as opposed to the traditional RE setting where a single sentence is input. Existing approaches rely on logical reasoning or contextual cues from entities. This paper reframes document-level RE as link prediction over a knowledge graph with distinct benefits: 1) Our approach combines entity context with document-derived logical reasoning, enhancing link prediction quality. 2) Predicted links between entities offer interpretability, elucidating employed reasoning. We evaluate our approach on three benchmark datasets: DocRED, ReDocRED, and DWIE. The results indicate that our proposed method outperforms the state-of-the-art models and suggests that incorporating context-based link prediction techniques can enhance the performance of document-level relation extraction models.
ReOnto: A Neuro-Symbolic Approach for Biomedical Relation Extraction
Sep 04, 2023Abstract:Relation Extraction (RE) is the task of extracting semantic relationships between entities in a sentence and aligning them to relations defined in a vocabulary, which is generally in the form of a Knowledge Graph (KG) or an ontology. Various approaches have been proposed so far to address this task. However, applying these techniques to biomedical text often yields unsatisfactory results because it is hard to infer relations directly from sentences due to the nature of the biomedical relations. To address these issues, we present a novel technique called ReOnto, that makes use of neuro symbolic knowledge for the RE task. ReOnto employs a graph neural network to acquire the sentence representation and leverages publicly accessible ontologies as prior knowledge to identify the sentential relation between two entities. The approach involves extracting the relation path between the two entities from the ontology. We evaluate the effect of using symbolic knowledge from ontologies with graph neural networks. Experimental results on two public biomedical datasets, BioRel and ADE, show that our method outperforms all the baselines (approximately by 3\%).
An Analysis of the Methods Employed for Breast Cancer Diagnosis
Jun 17, 2012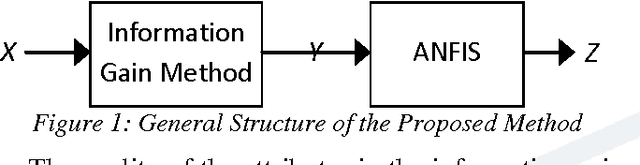
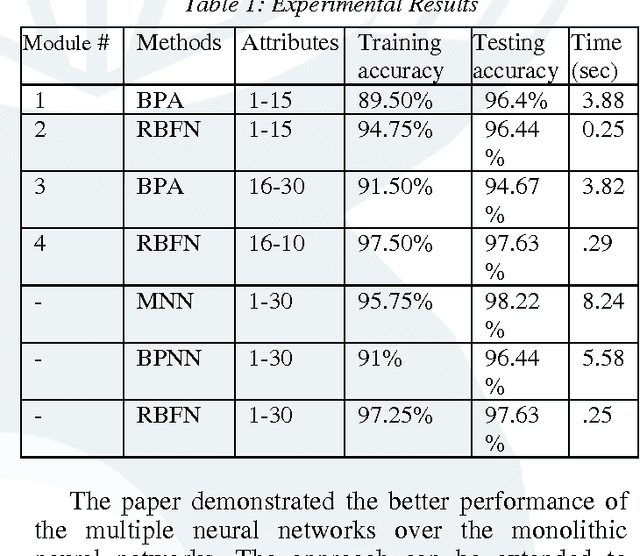
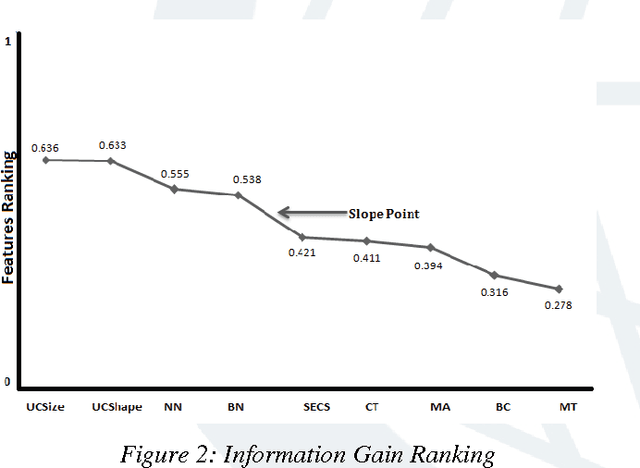
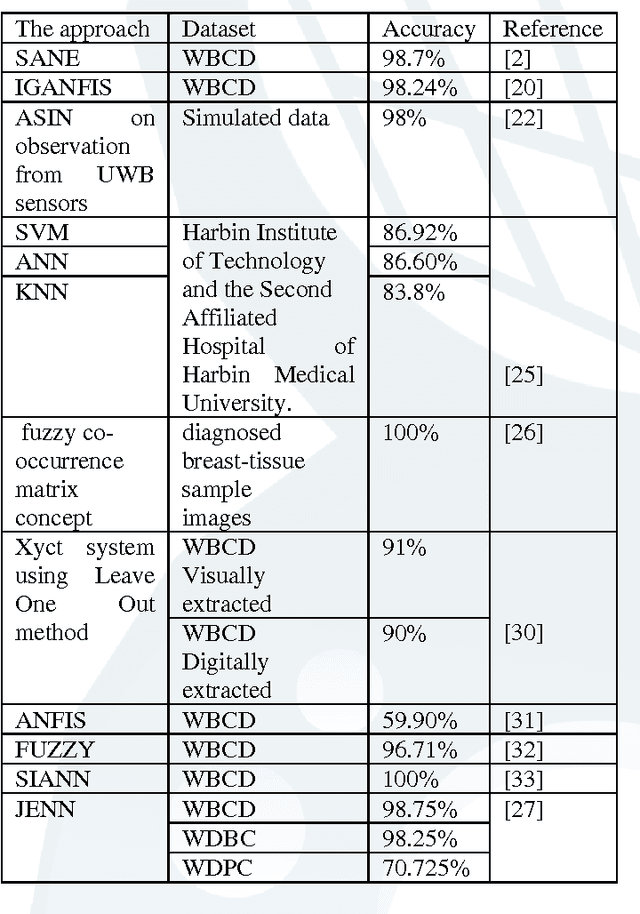
Abstract:Breast cancer research over the last decade has been tremendous. The ground breaking innovations and novel methods help in the early detection, in setting the stages of the therapy and in assessing the response of the patient to the treatment. The prediction of the recurrent cancer is also crucial for the survival of the patient. This paper studies various techniques used for the diagnosis of breast cancer. Different methods are explored for their merits and de-merits for the diagnosis of breast lesion. Some of the methods are yet unproven but the studies look very encouraging. It was found that the recent use of the combination of Artificial Neural Networks in most of the instances gives accurate results for the diagnosis of breast cancer and their use can also be extended to other diseases.
* 5 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge