Hongkuan Shi
IBoxCLA: Towards Robust Box-supervised Segmentation of Polyp via Improved Box-dice and Contrastive Latent-anchors
Oct 14, 2023



Abstract:Box-supervised polyp segmentation attracts increasing attention for its cost-effective potential. Existing solutions often rely on learning-free methods or pretrained models to laboriously generate pseudo masks, triggering Dice constraint subsequently. In this paper, we found that a model guided by the simplest box-filled masks can accurately predict polyp locations/sizes, but suffers from shape collapsing. In response, we propose two innovative learning fashions, Improved Box-dice (IBox) and Contrastive Latent-Anchors (CLA), and combine them to train a robust box-supervised model IBoxCLA. The core idea behind IBoxCLA is to decouple the learning of location/size and shape, allowing for focused constraints on each of them. Specifically, IBox transforms the segmentation map into a proxy map using shape decoupling and confusion-region swapping sequentially. Within the proxy map, shapes are disentangled, while locations/sizes are encoded as box-like responses. By constraining the proxy map instead of the raw prediction, the box-filled mask can well supervise IBoxCLA without misleading its shape learning. Furthermore, CLA contributes to shape learning by generating two types of latent anchors, which are learned and updated using momentum and segmented polyps to steadily represent polyp and background features. The latent anchors facilitate IBoxCLA to capture discriminative features within and outside boxes in a contrastive manner, yielding clearer boundaries. We benchmark IBoxCLA on five public polyp datasets. The experimental results demonstrate the competitive performance of IBoxCLA compared to recent fully-supervised polyp segmentation methods, and its superiority over other box-supervised state-of-the-arts with a relative increase of overall mDice and mIoU by at least 6.5% and 7.5%, respectively.
Bidirectional Semi-supervised Dual-branch CNN for Robust 3D Reconstruction of Stereo Endoscopic Images via Adaptive Cross and Parallel Supervisions
Oct 19, 2022
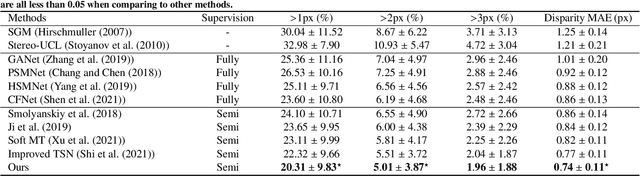

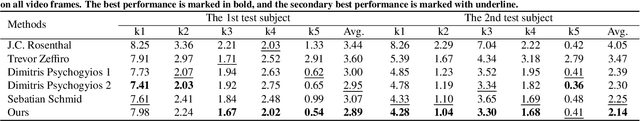
Abstract:Semi-supervised learning via teacher-student network can train a model effectively on a few labeled samples. It enables a student model to distill knowledge from the teacher's predictions of extra unlabeled data. However, such knowledge flow is typically unidirectional, having the performance vulnerable to the quality of teacher model. In this paper, we seek to robust 3D reconstruction of stereo endoscopic images by proposing a novel fashion of bidirectional learning between two learners, each of which can play both roles of teacher and student concurrently. Specifically, we introduce two self-supervisions, i.e., Adaptive Cross Supervision (ACS) and Adaptive Parallel Supervision (APS), to learn a dual-branch convolutional neural network. The two branches predict two different disparity probability distributions for the same position, and output their expectations as disparity values. The learned knowledge flows across branches along two directions: a cross direction (disparity guides distribution in ACS) and a parallel direction (disparity guides disparity in APS). Moreover, each branch also learns confidences to dynamically refine its provided supervisions. In ACS, the predicted disparity is softened into a unimodal distribution, and the lower the confidence, the smoother the distribution. In APS, the incorrect predictions are suppressed by lowering the weights of those with low confidence. With the adaptive bidirectional learning, the two branches enjoy well-tuned supervisions from each other, and eventually converge on a consistent and more accurate disparity estimation. The extensive and comprehensive experimental results on three public datasets demonstrate our superior performance over the fully-supervised and semi-supervised state-of-the-arts with a decrease of averaged disparity error by 13.95% and 3.90% at least, respectively.
Joint Progressive and Coarse-to-fine Registration of Brain MRI via Deformation Field Integration and Non-Rigid Feature Fusion
Sep 25, 2021



Abstract:Registration of brain MRI images requires to solve a deformation field, which is extremely difficult in aligning intricate brain tissues, e.g., subcortical nuclei, etc. Existing efforts resort to decomposing the target deformation field into intermediate sub-fields with either tiny motions, i.e., progressive registration stage by stage, or lower resolutions, i.e., coarse-to-fine estimation of the full-size deformation field. In this paper, we argue that those efforts are not mutually exclusive, and propose a unified framework for robust brain MRI registration in both progressive and coarse-to-fine manners simultaneously. Specifically, building on a dual-encoder U-Net, the fixed-moving MRI pair is encoded and decoded into multi-scale deformation sub-fields from coarse to fine. Each decoding block contains two proposed novel modules: i) in Deformation Field Integration (DFI), a single integrated sub-field is calculated, warping by which is equivalent to warping progressively by sub-fields from all previous decoding blocks, and ii) in Non-rigid Feature Fusion (NFF), features of the fixed-moving pair are aligned by DFI-integrated sub-field, and then fused to predict a finer sub-field. Leveraging both DFI and NFF, the target deformation field is factorized into multi-scale sub-fields, where the coarser fields alleviate the estimate of a finer one and the finer field learns to make up those misalignments insolvable by previous coarser ones. The extensive and comprehensive experimental results on both private and public datasets demonstrate a superior registration performance of brain MRI images over progressive registration only and coarse-to-fine estimation only, with an increase by at most 10% in the average Dice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge