Sunyang Fu
N3C Natural Language Processing
Digital Twin AI: Opportunities and Challenges from Large Language Models to World Models
Jan 04, 2026Abstract:Digital twins, as precise digital representations of physical systems, have evolved from passive simulation tools into intelligent and autonomous entities through the integration of artificial intelligence technologies. This paper presents a unified four-stage framework that systematically characterizes AI integration across the digital twin lifecycle, spanning modeling, mirroring, intervention, and autonomous management. By synthesizing existing technologies and practices, we distill a unified four-stage framework that systematically characterizes how AI methodologies are embedded across the digital twin lifecycle: (1) modeling the physical twin through physics-based and physics-informed AI approaches, (2) mirroring the physical system into a digital twin with real-time synchronization, (3) intervening in the physical twin through predictive modeling, anomaly detection, and optimization strategies, and (4) achieving autonomous management through large language models, foundation models, and intelligent agents. We analyze the synergy between physics-based modeling and data-driven learning, highlighting the shift from traditional numerical solvers to physics-informed and foundation models for physical systems. Furthermore, we examine how generative AI technologies, including large language models and generative world models, transform digital twins into proactive and self-improving cognitive systems capable of reasoning, communication, and creative scenario generation. Through a cross-domain review spanning eleven application domains, including healthcare, aerospace, smart manufacturing, robotics, and smart cities, we identify common challenges related to scalability, explainability, and trustworthiness, and outline directions for responsible AI-driven digital twin systems.
An Agentic Model Context Protocol Framework for Medical Concept Standardization
Sep 04, 2025Abstract:The Observational Medical Outcomes Partnership (OMOP) common data model (CDM) provides a standardized representation of heterogeneous health data to support large-scale, multi-institutional research. One critical step in data standardization using OMOP CDM is the mapping of source medical terms to OMOP standard concepts, a procedure that is resource-intensive and error-prone. While large language models (LLMs) have the potential to facilitate this process, their tendency toward hallucination makes them unsuitable for clinical deployment without training and expert validation. Here, we developed a zero-training, hallucination-preventive mapping system based on the Model Context Protocol (MCP), a standardized and secure framework allowing LLMs to interact with external resources and tools. The system enables explainable mapping and significantly improves efficiency and accuracy with minimal effort. It provides real-time vocabulary lookups and structured reasoning outputs suitable for immediate use in both exploratory and production environments.
A Literature Review and Framework for Human Evaluation of Generative Large Language Models in Healthcare
May 04, 2024Abstract:As generative artificial intelligence (AI), particularly Large Language Models (LLMs), continues to permeate healthcare, it remains crucial to supplement traditional automated evaluations with human expert evaluation. Understanding and evaluating the generated texts is vital for ensuring safety, reliability, and effectiveness. However, the cumbersome, time-consuming, and non-standardized nature of human evaluation presents significant obstacles to the widespread adoption of LLMs in practice. This study reviews existing literature on human evaluation methodologies for LLMs within healthcare. We highlight a notable need for a standardized and consistent human evaluation approach. Our extensive literature search, adhering to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, spans publications from January 2018 to February 2024. This review provides a comprehensive overview of the human evaluation approaches used in diverse healthcare applications.This analysis examines the human evaluation of LLMs across various medical specialties, addressing factors such as evaluation dimensions, sample types, and sizes, the selection and recruitment of evaluators, frameworks and metrics, the evaluation process, and statistical analysis of the results. Drawing from diverse evaluation strategies highlighted in these studies, we propose a comprehensive and practical framework for human evaluation of generative LLMs, named QUEST: Quality of Information, Understanding and Reasoning, Expression Style and Persona, Safety and Harm, and Trust and Confidence. This framework aims to improve the reliability, generalizability, and applicability of human evaluation of generative LLMs in different healthcare applications by defining clear evaluation dimensions and offering detailed guidelines.
BiomedGPT: A Unified and Generalist Biomedical Generative Pre-trained Transformer for Vision, Language, and Multimodal Tasks
May 26, 2023Abstract:In this paper, we introduce a unified and generalist Biomedical Generative Pre-trained Transformer (BiomedGPT) model, which leverages self-supervision on large and diverse datasets to accept multi-modal inputs and perform a range of downstream tasks. Our experiments demonstrate that BiomedGPT delivers expansive and inclusive representations of biomedical data, outperforming the majority of preceding state-of-the-art models across five distinct tasks with 20 public datasets spanning over 15 unique biomedical modalities. Through the ablation study, we also showcase the efficacy of our multi-modal and multi-task pretraining approach in transferring knowledge to previously unseen data. Overall, our work presents a significant step forward in developing unified and generalist models for biomedicine, with far-reaching implications for improving healthcare outcomes.
Detecting Reddit Users with Depression Using a Hybrid Neural Network
Feb 03, 2023



Abstract:Depression is a widespread mental health issue, affecting an estimated 3.8% of the global population. It is also one of the main contributors to disability worldwide. Recently it is becoming popular for individuals to use social media platforms (e.g., Reddit) to express their difficulties and health issues (e.g., depression) and seek support from other users in online communities. It opens great opportunities to automatically identify social media users with depression by parsing millions of posts for potential interventions. Deep learning methods have begun to dominate in the field of machine learning and natural language processing (NLP) because of their ease of use, efficient processing, and state-of-the-art results on many NLP tasks. In this work, we propose a hybrid deep learning model which combines a pretrained sentence BERT (SBERT) and convolutional neural network (CNN) to detect individuals with depression with their Reddit posts. The sentence BERT is used to learn the meaningful representation of semantic information in each post. CNN enables the further transformation of those embeddings and the temporal identification of behavioral patterns of users. We trained and evaluated the model performance to identify Reddit users with depression by utilizing the Self-reported Mental Health Diagnoses (SMHD) data. The hybrid deep learning model achieved an accuracy of 0.86 and an F1 score of 0.86 and outperformed the state-of-the-art documented result (F1 score of 0.79) by other machine learning models in the literature. The results show the feasibility of the hybrid model to identify individuals with depression. Although the hybrid model is validated to detect depression with Reddit posts, it can be easily tuned and applied to other text classification tasks and different clinical applications.
An Open Natural Language Processing Development Framework for EHR-based Clinical Research: A case demonstration using the National COVID Cohort Collaborative (N3C)
Oct 20, 2021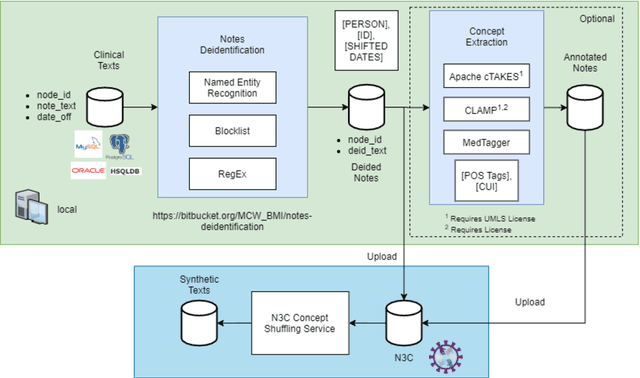
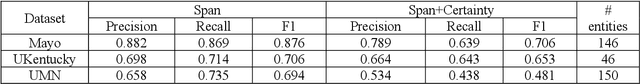
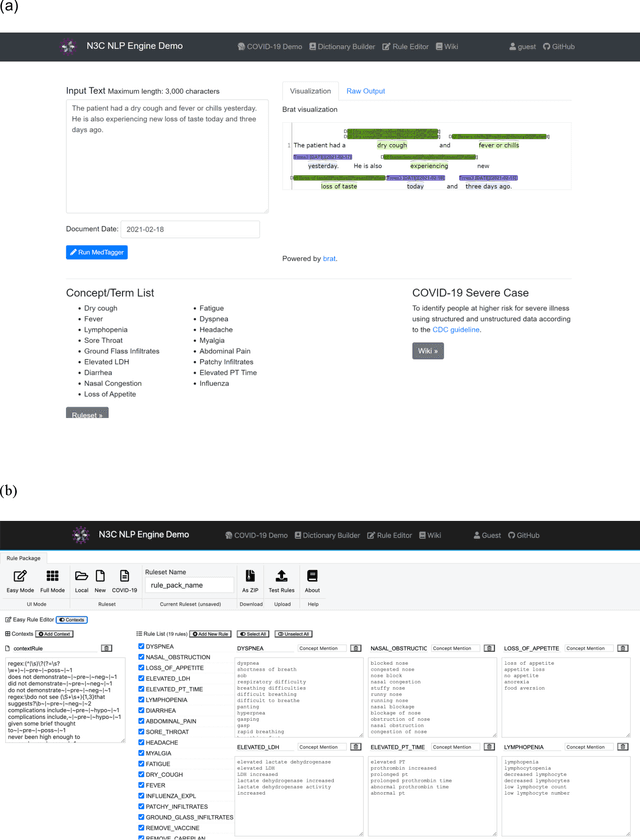
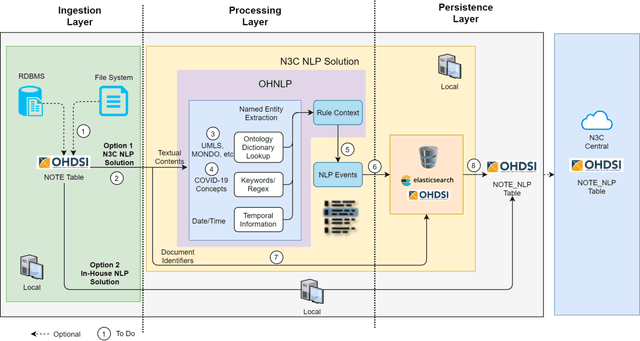
Abstract:While we pay attention to the latest advances in clinical natural language processing (NLP), we can notice some resistance in the clinical and translational research community to adopt NLP models due to limited transparency, Interpretability and usability. Built upon our previous work, in this study, we proposed an open natural language processing development framework and evaluated it through the implementation of NLP algorithms for the National COVID Cohort Collaborative (N3C). Based on the interests in information extraction from COVID-19 related clinical notes, our work includes 1) an open data annotation process using COVID-19 signs and symptoms as the use case, 2) a community-driven ruleset composing platform, and 3) a synthetic text data generation workflow to generate texts for information extraction tasks without involving human subjects. The generated corpora derived out of the texts from multiple intuitions and gold standard annotation are tested on a single institution's rule set has the performances in F1 score of 0.876, 0.706 and 0.694, respectively. The study as a consortium effort of the N3C NLP subgroup demonstrates the feasibility of creating a federated NLP algorithm development and benchmarking platform to enhance multi-institution clinical NLP study.
Neural Language Models with Distant Supervision to Identify Major Depressive Disorder from Clinical Notes
Apr 19, 2021
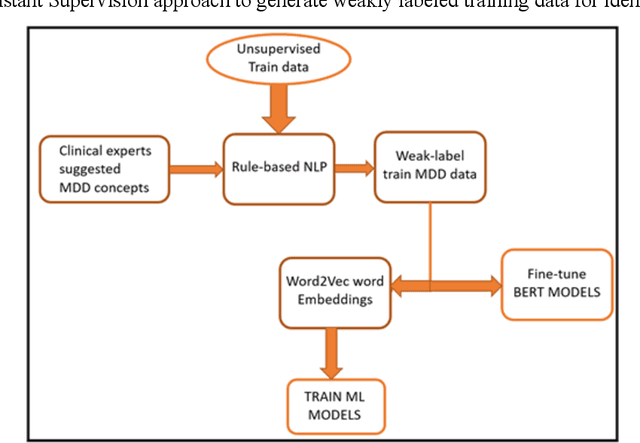
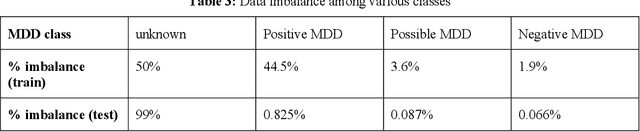
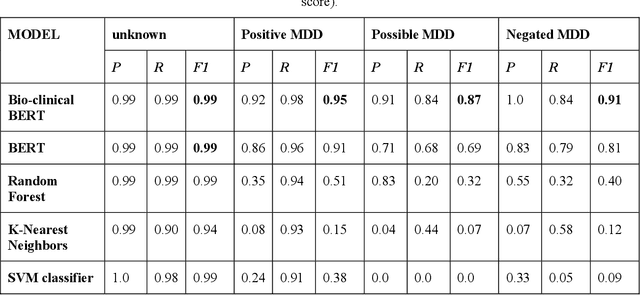
Abstract:Major depressive disorder (MDD) is a prevalent psychiatric disorder that is associated with significant healthcare burden worldwide. Phenotyping of MDD can help early diagnosis and consequently may have significant advantages in patient management. In prior research MDD phenotypes have been extracted from structured Electronic Health Records (EHR) or using Electroencephalographic (EEG) data with traditional machine learning models to predict MDD phenotypes. However, MDD phenotypic information is also documented in free-text EHR data, such as clinical notes. While clinical notes may provide more accurate phenotyping information, natural language processing (NLP) algorithms must be developed to abstract such information. Recent advancements in NLP resulted in state-of-the-art neural language models, such as Bidirectional Encoder Representations for Transformers (BERT) model, which is a transformer-based model that can be pre-trained from a corpus of unsupervised text data and then fine-tuned on specific tasks. However, such neural language models have been underutilized in clinical NLP tasks due to the lack of large training datasets. In the literature, researchers have utilized the distant supervision paradigm to train machine learning models on clinical text classification tasks to mitigate the issue of lacking annotated training data. It is still unknown whether the paradigm is effective for neural language models. In this paper, we propose to leverage the neural language models in a distant supervision paradigm to identify MDD phenotypes from clinical notes. The experimental results indicate that our proposed approach is effective in identifying MDD phenotypes and that the Bio- Clinical BERT, a specific BERT model for clinical data, achieved the best performance in comparison with conventional machine learning models.
Development of Clinical Concept Extraction Applications: A Methodology Review
Oct 28, 2019
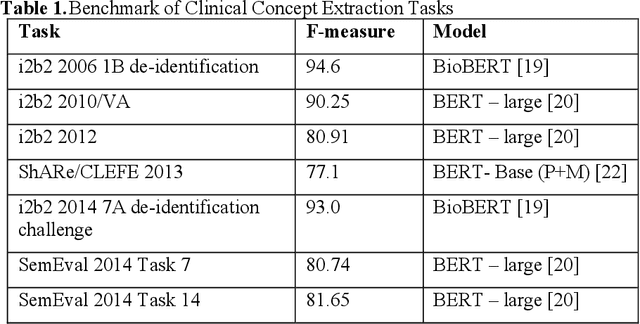
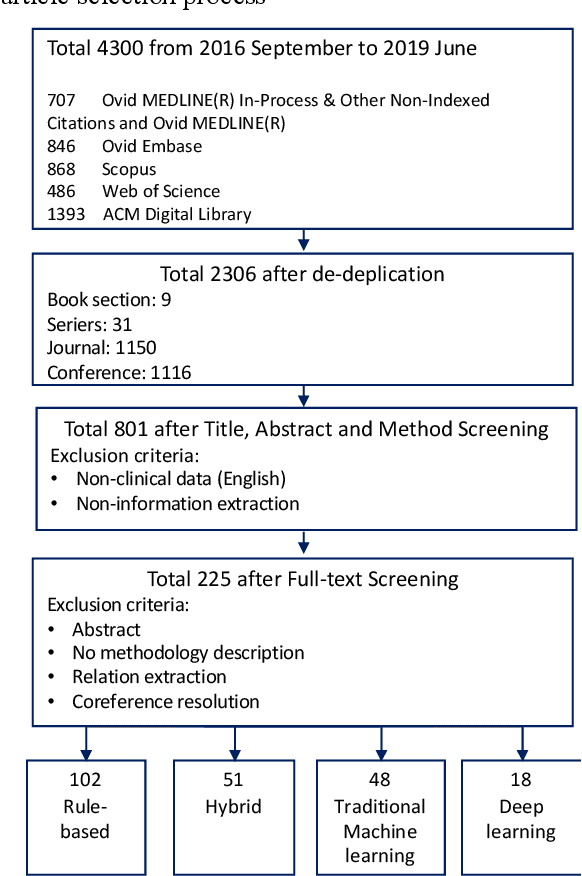
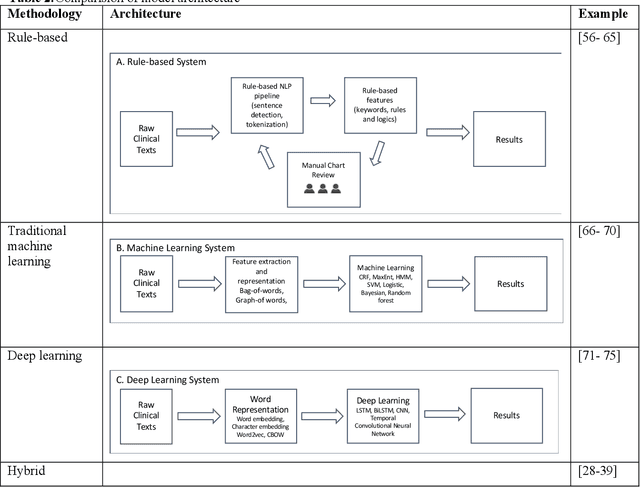
Abstract:Our study provided a review of the development of clinical concept extraction applications from January 2009 to June 2019. We hope, through the studying of different approaches with variant clinical context, can enhance the decision making for the development of clinical concept extraction.
How Good is Artificial Intelligence at Automatically Answering Consumer Questions Related to Alzheimer's Disease?
Aug 21, 2019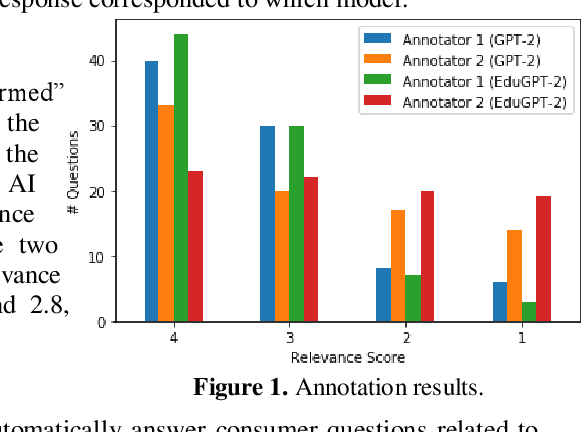
Abstract:Alzheimer's Disease (AD) is the most common type of dementia, comprising 60-80% of cases. There were an estimated 5.8 million Americans living with Alzheimer's dementia in 2019, and this number will almost double every 20 years. The total lifetime cost of care for someone with dementia is estimated to be $350,174 in 2018, 70% of which is associated with family-provided care. Most family caregivers face emotional, financial and physical difficulties. As a medium to relieve this burden, online communities in social media websites such as Twitter, Reddit, and Yahoo! Answers provide potential venues for caregivers to search relevant questions and answers, or post questions and seek answers from other members. However, there are often a limited number of relevant questions and responses to search from, and posted questions are rarely answered immediately. Due to recent advancement in Artificial Intelligence (AI), particularly Natural Language Processing (NLP), we propose to utilize AI to automatically generate answers to AD-related consumer questions posted by caregivers and evaluate how good AI is at answering those questions. To the best of our knowledge, this is the first study in the literature applying and evaluating AI models designed to automatically answer consumer questions related to AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge