Hongfang Liu
N3C Natural Language Processing
Toward Global Large Language Models in Medicine
Jan 05, 2026Abstract:Despite continuous advances in medical technology, the global distribution of health care resources remains uneven. The development of large language models (LLMs) has transformed the landscape of medicine and holds promise for improving health care quality and expanding access to medical information globally. However, existing LLMs are primarily trained on high-resource languages, limiting their applicability in global medical scenarios. To address this gap, we constructed GlobMed, a large multilingual medical dataset, containing over 500,000 entries spanning 12 languages, including four low-resource languages. Building on this, we established GlobMed-Bench, which systematically assesses 56 state-of-the-art proprietary and open-weight LLMs across multiple multilingual medical tasks, revealing significant performance disparities across languages, particularly for low-resource languages. Additionally, we introduced GlobMed-LLMs, a suite of multilingual medical LLMs trained on GlobMed, with parameters ranging from 1.7B to 8B. GlobMed-LLMs achieved an average performance improvement of over 40% relative to baseline models, with a more than threefold increase in performance on low-resource languages. Together, these resources provide an important foundation for advancing the equitable development and application of LLMs globally, enabling broader language communities to benefit from technological advances.
An Agentic Model Context Protocol Framework for Medical Concept Standardization
Sep 04, 2025Abstract:The Observational Medical Outcomes Partnership (OMOP) common data model (CDM) provides a standardized representation of heterogeneous health data to support large-scale, multi-institutional research. One critical step in data standardization using OMOP CDM is the mapping of source medical terms to OMOP standard concepts, a procedure that is resource-intensive and error-prone. While large language models (LLMs) have the potential to facilitate this process, their tendency toward hallucination makes them unsuitable for clinical deployment without training and expert validation. Here, we developed a zero-training, hallucination-preventive mapping system based on the Model Context Protocol (MCP), a standardized and secure framework allowing LLMs to interact with external resources and tools. The system enables explainable mapping and significantly improves efficiency and accuracy with minimal effort. It provides real-time vocabulary lookups and structured reasoning outputs suitable for immediate use in both exploratory and production environments.
Multi-Label Classification with Generative AI Models in Healthcare: A Case Study of Suicidality and Risk Factors
Jul 22, 2025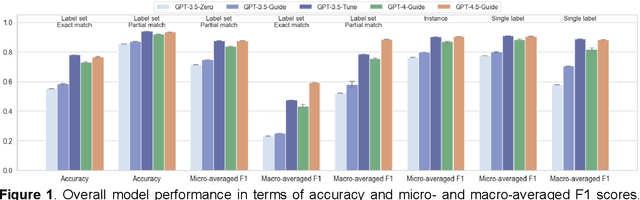
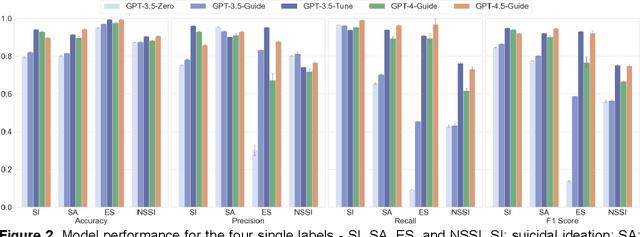
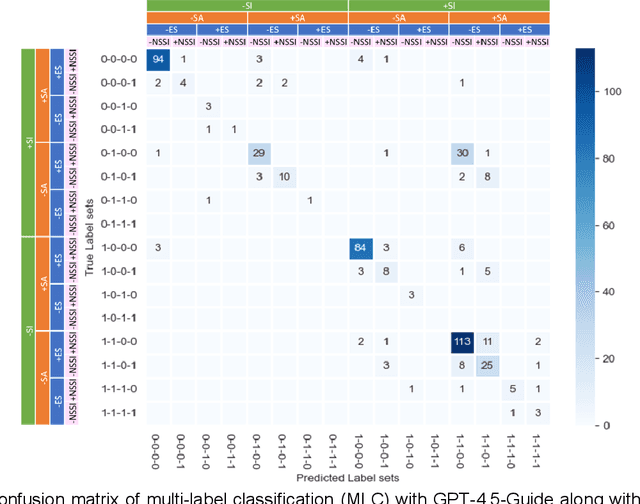
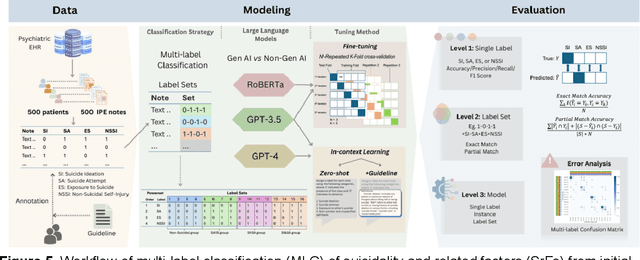
Abstract:Suicide remains a pressing global health crisis, with over 720,000 deaths annually and millions more affected by suicide ideation (SI) and suicide attempts (SA). Early identification of suicidality-related factors (SrFs), including SI, SA, exposure to suicide (ES), and non-suicidal self-injury (NSSI), is critical for timely intervention. While prior studies have applied AI to detect SrFs in clinical notes, most treat suicidality as a binary classification task, overlooking the complexity of cooccurring risk factors. This study explores the use of generative large language models (LLMs), specifically GPT-3.5 and GPT-4.5, for multi-label classification (MLC) of SrFs from psychiatric electronic health records (EHRs). We present a novel end to end generative MLC pipeline and introduce advanced evaluation methods, including label set level metrics and a multilabel confusion matrix for error analysis. Finetuned GPT-3.5 achieved top performance with 0.94 partial match accuracy and 0.91 F1 score, while GPT-4.5 with guided prompting showed superior performance across label sets, including rare or minority label sets, indicating a more balanced and robust performance. Our findings reveal systematic error patterns, such as the conflation of SI and SA, and highlight the models tendency toward cautious over labeling. This work not only demonstrates the feasibility of using generative AI for complex clinical classification tasks but also provides a blueprint for structuring unstructured EHR data to support large scale clinical research and evidence based medicine.
Scaling Up Biomedical Vision-Language Models: Fine-Tuning, Instruction Tuning, and Multi-Modal Learning
May 23, 2025Abstract:To advance biomedical vison-language model capabilities through scaling up, fine-tuning, and instruction tuning, develop vision-language models with improved performance in handling long text, explore strategies to efficiently adopt vision language models for diverse multi-modal biomedical tasks, and examine the zero-shot learning performance. We developed two biomedical vision language models, BiomedGPT-Large and BiomedGPT-XLarge, based on an encoder-decoder-based transformer architecture. We fine-tuned the two models on 23 benchmark datasets from 6 multi-modal biomedical tasks including one image-only task (image classification), three language-only tasks (text understanding, text summarization and question answering), and two vision-language tasks (visual question answering and image captioning). We compared the developed scaled models with our previous BiomedGPT-Base model and existing prestigious models reported in the literature. We instruction-tuned the two models using a large-scale multi-modal biomedical instruction-tuning dataset and assessed the zero-shot learning performance and alignment accuracy.
Explainable Diagnosis Prediction through Neuro-Symbolic Integration
Oct 01, 2024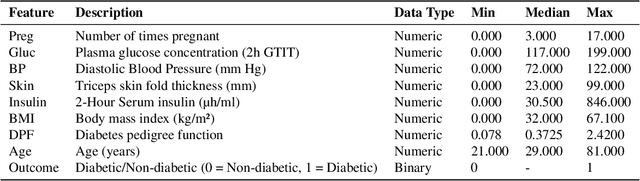

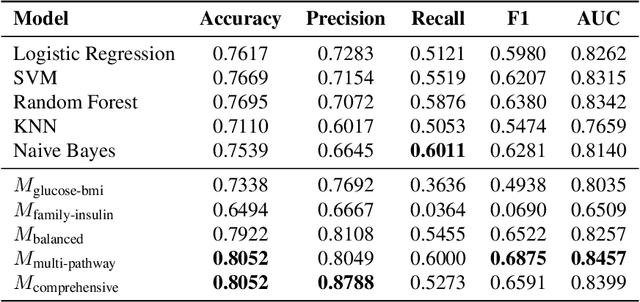
Abstract:Diagnosis prediction is a critical task in healthcare, where timely and accurate identification of medical conditions can significantly impact patient outcomes. Traditional machine learning and deep learning models have achieved notable success in this domain but often lack interpretability which is a crucial requirement in clinical settings. In this study, we explore the use of neuro-symbolic methods, specifically Logical Neural Networks (LNNs), to develop explainable models for diagnosis prediction. Essentially, we design and implement LNN-based models that integrate domain-specific knowledge through logical rules with learnable thresholds. Our models, particularly $M_{\text{multi-pathway}}$ and $M_{\text{comprehensive}}$, demonstrate superior performance over traditional models such as Logistic Regression, SVM, and Random Forest, achieving higher accuracy (up to 80.52\%) and AUROC scores (up to 0.8457) in the case study of diabetes prediction. The learned weights and thresholds within the LNN models provide direct insights into feature contributions, enhancing interpretability without compromising predictive power. These findings highlight the potential of neuro-symbolic approaches in bridging the gap between accuracy and explainability in healthcare AI applications. By offering transparent and adaptable diagnostic models, our work contributes to the advancement of precision medicine and supports the development of equitable healthcare solutions. Future research will focus on extending these methods to larger and more diverse datasets to further validate their applicability across different medical conditions and populations.
Suicide Phenotyping from Clinical Notes in Safety-Net Psychiatric Hospital Using Multi-Label Classification with Pre-Trained Language Models
Sep 27, 2024



Abstract:Accurate identification and categorization of suicidal events can yield better suicide precautions, reducing operational burden, and improving care quality in high-acuity psychiatric settings. Pre-trained language models offer promise for identifying suicidality from unstructured clinical narratives. We evaluated the performance of four BERT-based models using two fine-tuning strategies (multiple single-label and single multi-label) for detecting coexisting suicidal events from 500 annotated psychiatric evaluation notes. The notes were labeled for suicidal ideation (SI), suicide attempts (SA), exposure to suicide (ES), and non-suicidal self-injury (NSSI). RoBERTa outperformed other models using binary relevance (acc=0.86, F1=0.78). MentalBERT (F1=0.74) also exceeded BioClinicalBERT (F1=0.72). RoBERTa fine-tuned with a single multi-label classifier further improved performance (acc=0.88, F1=0.81), highlighting that models pre-trained on domain-relevant data and the single multi-label classification strategy enhance efficiency and performance. Keywords: EHR-based Phynotyping; Natural Language Processing; Secondary Use of EHR Data; Suicide Classification; BERT-based Model; Psychiatry; Mental Health
Large Language Models Struggle in Token-Level Clinical Named Entity Recognition
Jun 30, 2024Abstract:Large Language Models (LLMs) have revolutionized various sectors, including healthcare where they are employed in diverse applications. Their utility is particularly significant in the context of rare diseases, where data scarcity, complexity, and specificity pose considerable challenges. In the clinical domain, Named Entity Recognition (NER) stands out as an essential task and it plays a crucial role in extracting relevant information from clinical texts. Despite the promise of LLMs, current research mostly concentrates on document-level NER, identifying entities in a more general context across entire documents, without extracting their precise location. Additionally, efforts have been directed towards adapting ChatGPT for token-level NER. However, there is a significant research gap when it comes to employing token-level NER for clinical texts, especially with the use of local open-source LLMs. This study aims to bridge this gap by investigating the effectiveness of both proprietary and local LLMs in token-level clinical NER. Essentially, we delve into the capabilities of these models through a series of experiments involving zero-shot prompting, few-shot prompting, retrieval-augmented generation (RAG), and instruction-fine-tuning. Our exploration reveals the inherent challenges LLMs face in token-level NER, particularly in the context of rare diseases, and suggests possible improvements for their application in healthcare. This research contributes to narrowing a significant gap in healthcare informatics and offers insights that could lead to a more refined application of LLMs in the healthcare sector.
CancerLLM: A Large Language Model in Cancer Domain
Jun 15, 2024



Abstract:Medical Large Language Models (LLMs) such as ClinicalCamel 70B, Llama3-OpenBioLLM 70B have demonstrated impressive performance on a wide variety of medical NLP task.However, there still lacks a large language model (LLM) specifically designed for cancer domain. Moreover, these LLMs typically have billions of parameters, making them computationally expensive for healthcare systems.Thus, in this study, we propose CancerLLM, a model with 7 billion parameters and a Mistral-style architecture, pre-trained on 2,676,642 clinical notes and 515,524 pathology reports covering 17 cancer types, followed by fine-tuning on three cancer-relevant tasks, including cancer phenotypes extraction, cancer diagnosis generation, and cancer treatment plan generation. Our evaluation demonstrated that CancerLLM achieves state-of-the-art results compared to other existing LLMs, with an average F1 score improvement of 8.1\%. Additionally, CancerLLM outperforms other models on two proposed robustness testbeds. This illustrates that CancerLLM can be effectively applied to clinical AI systems, enhancing clinical research and healthcare delivery in the field of cancer.
GRU-D-Weibull: A Novel Real-Time Individualized Endpoint Prediction
Aug 14, 2023



Abstract:Accurate prediction models for individual-level endpoints and time-to-endpoints are crucial in clinical practice. In this study, we propose a novel approach, GRU-D-Weibull, which combines gated recurrent units with decay (GRU-D) to model the Weibull distribution. Our method enables real-time individualized endpoint prediction and population-level risk management. Using a cohort of 6,879 patients with stage 4 chronic kidney disease (CKD4), we evaluated the performance of GRU-D-Weibull in endpoint prediction. The C-index of GRU-D-Weibull was ~0.7 at the index date and increased to ~0.77 after 4.3 years of follow-up, similar to random survival forest. Our approach achieved an absolute L1-loss of ~1.1 years (SD 0.95) at the CKD4 index date and a minimum of ~0.45 years (SD0.3) at 4 years of follow-up, outperforming competing methods significantly. GRU-D-Weibull consistently constrained the predicted survival probability at the time of an event within a smaller and more fixed range compared to other models throughout the follow-up period. We observed significant correlations between the error in point estimates and missing proportions of input features at the index date (correlations from ~0.1 to ~0.3), which diminished within 1 year as more data became available. By post-training recalibration, we successfully aligned the predicted and observed survival probabilities across multiple prediction horizons at different time points during follow-up. Our findings demonstrate the considerable potential of GRU-D-Weibull as the next-generation architecture for endpoint risk management, capable of generating various endpoint estimates for real-time monitoring using clinical data.
BiomedGPT: A Unified and Generalist Biomedical Generative Pre-trained Transformer for Vision, Language, and Multimodal Tasks
May 26, 2023Abstract:In this paper, we introduce a unified and generalist Biomedical Generative Pre-trained Transformer (BiomedGPT) model, which leverages self-supervision on large and diverse datasets to accept multi-modal inputs and perform a range of downstream tasks. Our experiments demonstrate that BiomedGPT delivers expansive and inclusive representations of biomedical data, outperforming the majority of preceding state-of-the-art models across five distinct tasks with 20 public datasets spanning over 15 unique biomedical modalities. Through the ablation study, we also showcase the efficacy of our multi-modal and multi-task pretraining approach in transferring knowledge to previously unseen data. Overall, our work presents a significant step forward in developing unified and generalist models for biomedicine, with far-reaching implications for improving healthcare outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge