Qiuhao Lu
A Comprehensive Survey of Small Language Models in the Era of Large Language Models: Techniques, Enhancements, Applications, Collaboration with LLMs, and Trustworthiness
Nov 04, 2024Abstract:Large language models (LLM) have demonstrated emergent abilities in text generation, question answering, and reasoning, facilitating various tasks and domains. Despite their proficiency in various tasks, LLMs like LaPM 540B and Llama-3.1 405B face limitations due to large parameter sizes and computational demands, often requiring cloud API use which raises privacy concerns, limits real-time applications on edge devices, and increases fine-tuning costs. Additionally, LLMs often underperform in specialized domains such as healthcare and law due to insufficient domain-specific knowledge, necessitating specialized models. Therefore, Small Language Models (SLMs) are increasingly favored for their low inference latency, cost-effectiveness, efficient development, and easy customization and adaptability. These models are particularly well-suited for resource-limited environments and domain knowledge acquisition, addressing LLMs' challenges and proving ideal for applications that require localized data handling for privacy, minimal inference latency for efficiency, and domain knowledge acquisition through lightweight fine-tuning. The rising demand for SLMs has spurred extensive research and development. However, a comprehensive survey investigating issues related to the definition, acquisition, application, enhancement, and reliability of SLM remains lacking, prompting us to conduct a detailed survey on these topics. The definition of SLMs varies widely, thus to standardize, we propose defining SLMs by their capability to perform specialized tasks and suitability for resource-constrained settings, setting boundaries based on the minimal size for emergent abilities and the maximum size sustainable under resource constraints. For other aspects, we provide a taxonomy of relevant models/methods and develop general frameworks for each category to enhance and utilize SLMs effectively.
Explainable Diagnosis Prediction through Neuro-Symbolic Integration
Oct 01, 2024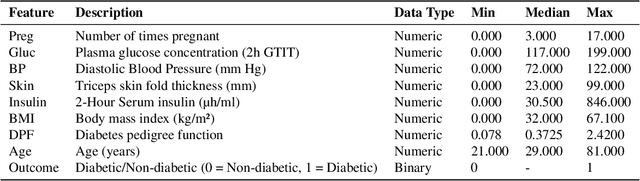

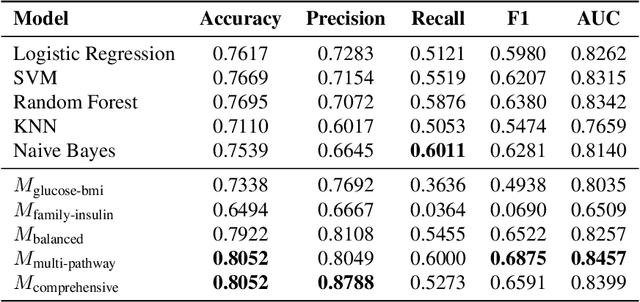
Abstract:Diagnosis prediction is a critical task in healthcare, where timely and accurate identification of medical conditions can significantly impact patient outcomes. Traditional machine learning and deep learning models have achieved notable success in this domain but often lack interpretability which is a crucial requirement in clinical settings. In this study, we explore the use of neuro-symbolic methods, specifically Logical Neural Networks (LNNs), to develop explainable models for diagnosis prediction. Essentially, we design and implement LNN-based models that integrate domain-specific knowledge through logical rules with learnable thresholds. Our models, particularly $M_{\text{multi-pathway}}$ and $M_{\text{comprehensive}}$, demonstrate superior performance over traditional models such as Logistic Regression, SVM, and Random Forest, achieving higher accuracy (up to 80.52\%) and AUROC scores (up to 0.8457) in the case study of diabetes prediction. The learned weights and thresholds within the LNN models provide direct insights into feature contributions, enhancing interpretability without compromising predictive power. These findings highlight the potential of neuro-symbolic approaches in bridging the gap between accuracy and explainability in healthcare AI applications. By offering transparent and adaptable diagnostic models, our work contributes to the advancement of precision medicine and supports the development of equitable healthcare solutions. Future research will focus on extending these methods to larger and more diverse datasets to further validate their applicability across different medical conditions and populations.
Large Language Models Struggle in Token-Level Clinical Named Entity Recognition
Jun 30, 2024Abstract:Large Language Models (LLMs) have revolutionized various sectors, including healthcare where they are employed in diverse applications. Their utility is particularly significant in the context of rare diseases, where data scarcity, complexity, and specificity pose considerable challenges. In the clinical domain, Named Entity Recognition (NER) stands out as an essential task and it plays a crucial role in extracting relevant information from clinical texts. Despite the promise of LLMs, current research mostly concentrates on document-level NER, identifying entities in a more general context across entire documents, without extracting their precise location. Additionally, efforts have been directed towards adapting ChatGPT for token-level NER. However, there is a significant research gap when it comes to employing token-level NER for clinical texts, especially with the use of local open-source LLMs. This study aims to bridge this gap by investigating the effectiveness of both proprietary and local LLMs in token-level clinical NER. Essentially, we delve into the capabilities of these models through a series of experiments involving zero-shot prompting, few-shot prompting, retrieval-augmented generation (RAG), and instruction-fine-tuning. Our exploration reveals the inherent challenges LLMs face in token-level NER, particularly in the context of rare diseases, and suggests possible improvements for their application in healthcare. This research contributes to narrowing a significant gap in healthcare informatics and offers insights that could lead to a more refined application of LLMs in the healthcare sector.
Textual Data Augmentation for Patient Outcomes Prediction
Nov 13, 2022



Abstract:Deep learning models have demonstrated superior performance in various healthcare applications. However, the major limitation of these deep models is usually the lack of high-quality training data due to the private and sensitive nature of this field. In this study, we propose a novel textual data augmentation method to generate artificial clinical notes in patients' Electronic Health Records (EHRs) that can be used as additional training data for patient outcomes prediction. Essentially, we fine-tune the generative language model GPT-2 to synthesize labeled text with the original training data. More specifically, We propose a teacher-student framework where we first pre-train a teacher model on the original data, and then train a student model on the GPT-augmented data under the guidance of the teacher. We evaluate our method on the most common patient outcome, i.e., the 30-day readmission rate. The experimental results show that deep models can improve their predictive performance with the augmented data, indicating the effectiveness of the proposed architecture.
Predicting Patient Readmission Risk from Medical Text via Knowledge Graph Enhanced Multiview Graph Convolution
Dec 19, 2021



Abstract:Unplanned intensive care unit (ICU) readmission rate is an important metric for evaluating the quality of hospital care. Efficient and accurate prediction of ICU readmission risk can not only help prevent patients from inappropriate discharge and potential dangers, but also reduce associated costs of healthcare. In this paper, we propose a new method that uses medical text of Electronic Health Records (EHRs) for prediction, which provides an alternative perspective to previous studies that heavily depend on numerical and time-series features of patients. More specifically, we extract discharge summaries of patients from their EHRs, and represent them with multiview graphs enhanced by an external knowledge graph. Graph convolutional networks are then used for representation learning. Experimental results prove the effectiveness of our method, yielding state-of-the-art performance for this task.
LNN-EL: A Neuro-Symbolic Approach to Short-text Entity Linking
Jun 17, 2021
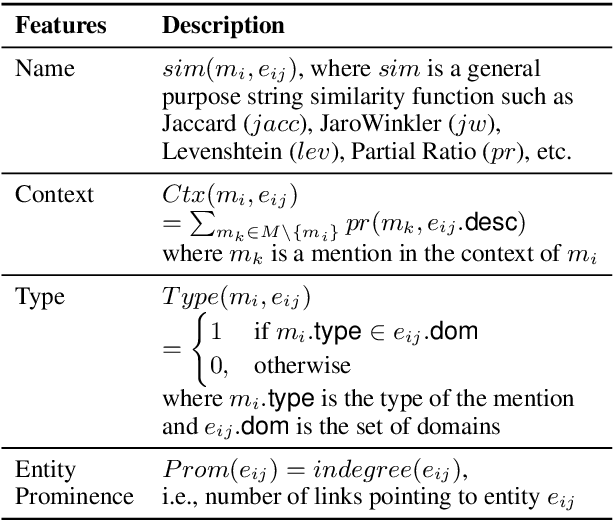
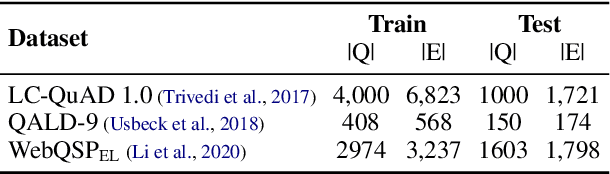
Abstract:Entity linking (EL), the task of disambiguating mentions in text by linking them to entities in a knowledge graph, is crucial for text understanding, question answering or conversational systems. Entity linking on short text (e.g., single sentence or question) poses particular challenges due to limited context. While prior approaches use either heuristics or black-box neural methods, here we propose LNN-EL, a neuro-symbolic approach that combines the advantages of using interpretable rules based on first-order logic with the performance of neural learning. Even though constrained to using rules, LNN-EL performs competitively against SotA black-box neural approaches, with the added benefits of extensibility and transferability. In particular, we show that we can easily blend existing rule templates given by a human expert, with multiple types of features (priors, BERT encodings, box embeddings, etc), and even scores resulting from previous EL methods, thus improving on such methods. For instance, on the LC-QuAD-1.0 dataset, we show more than $4$\% increase in F1 score over previous SotA. Finally, we show that the inductive bias offered by using logic results in learned rules that transfer well across datasets, even without fine tuning, while maintaining high accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge