Jungwei W. Fan
Artificial Intelligence-Enabled Analysis of Radiology Reports: Epidemiology and Consequences of Incidental Thyroid Findings
Oct 30, 2025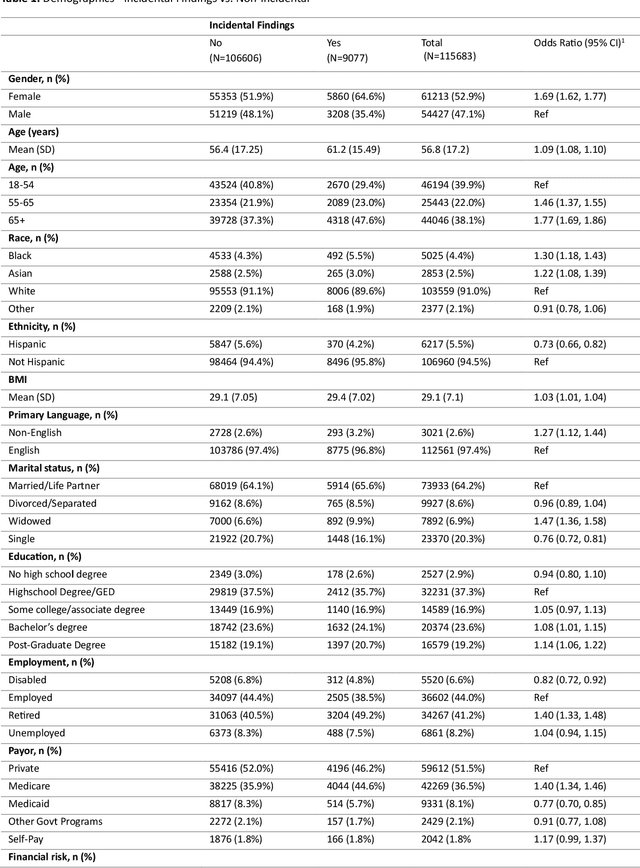
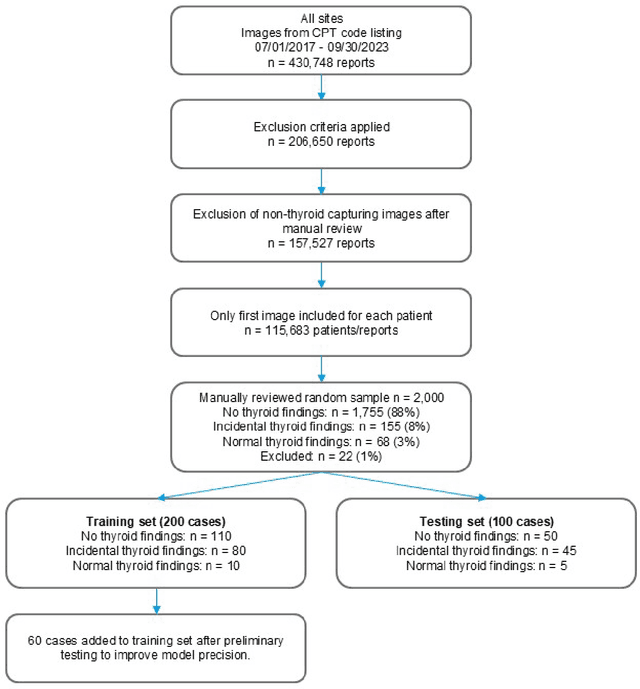
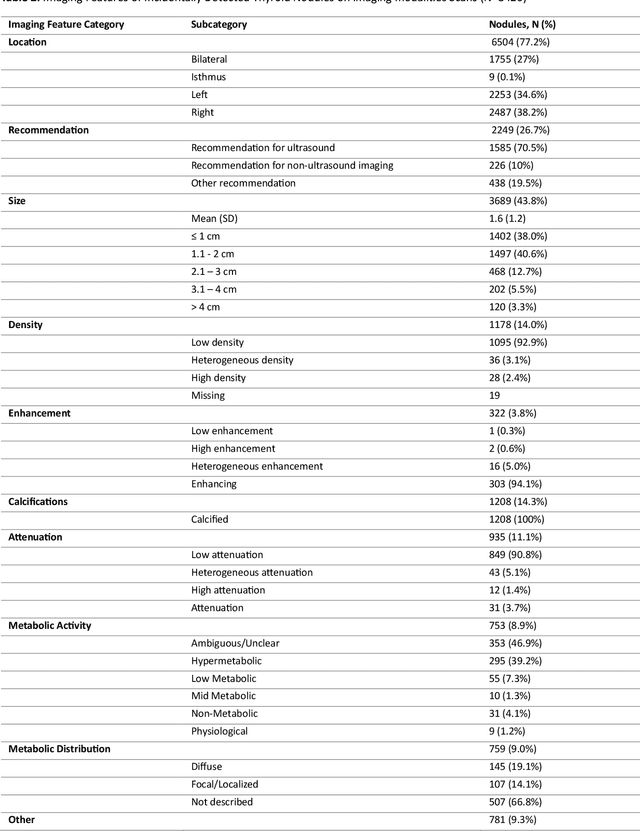
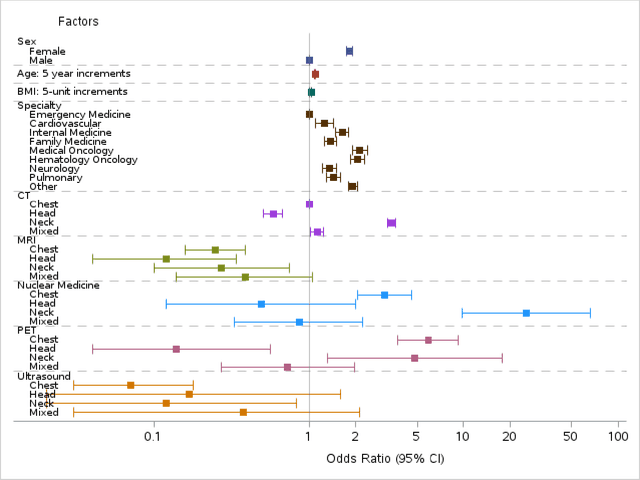
Abstract:Importance Incidental thyroid findings (ITFs) are increasingly detected on imaging performed for non-thyroid indications. Their prevalence, features, and clinical consequences remain undefined. Objective To develop, validate, and deploy a natural language processing (NLP) pipeline to identify ITFs in radiology reports and assess their prevalence, features, and clinical outcomes. Design, Setting, and Participants Retrospective cohort of adults without prior thyroid disease undergoing thyroid-capturing imaging at Mayo Clinic sites from July 1, 2017, to September 30, 2023. A transformer-based NLP pipeline identified ITFs and extracted nodule characteristics from image reports from multiple modalities and body regions. Main Outcomes and Measures Prevalence of ITFs, downstream thyroid ultrasound, biopsy, thyroidectomy, and thyroid cancer diagnosis. Logistic regression identified demographic and imaging-related factors. Results Among 115,683 patients (mean age, 56.8 [SD 17.2] years; 52.9% women), 9,077 (7.8%) had an ITF, of which 92.9% were nodules. ITFs were more likely in women, older adults, those with higher BMI, and when imaging was ordered by oncology or internal medicine. Compared with chest CT, ITFs were more likely via neck CT, PET, and nuclear medicine scans. Nodule characteristics were poorly documented, with size reported in 44% and other features in fewer than 15% (e.g. calcifications). Compared with patients without ITFs, those with ITFs had higher odds of thyroid nodule diagnosis, biopsy, thyroidectomy and thyroid cancer diagnosis. Most cancers were papillary, and larger when detected after ITFs vs no ITF. Conclusions ITFs were common and strongly associated with cascades leading to the detection of small, low-risk cancers. These findings underscore the role of ITFs in thyroid cancer overdiagnosis and the need for standardized reporting and more selective follow-up.
Explainable Diagnosis Prediction through Neuro-Symbolic Integration
Oct 01, 2024

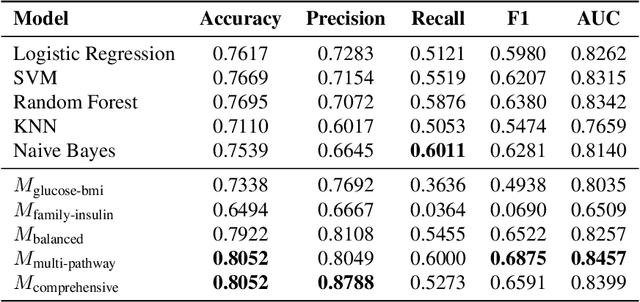
Abstract:Diagnosis prediction is a critical task in healthcare, where timely and accurate identification of medical conditions can significantly impact patient outcomes. Traditional machine learning and deep learning models have achieved notable success in this domain but often lack interpretability which is a crucial requirement in clinical settings. In this study, we explore the use of neuro-symbolic methods, specifically Logical Neural Networks (LNNs), to develop explainable models for diagnosis prediction. Essentially, we design and implement LNN-based models that integrate domain-specific knowledge through logical rules with learnable thresholds. Our models, particularly $M_{\text{multi-pathway}}$ and $M_{\text{comprehensive}}$, demonstrate superior performance over traditional models such as Logistic Regression, SVM, and Random Forest, achieving higher accuracy (up to 80.52\%) and AUROC scores (up to 0.8457) in the case study of diabetes prediction. The learned weights and thresholds within the LNN models provide direct insights into feature contributions, enhancing interpretability without compromising predictive power. These findings highlight the potential of neuro-symbolic approaches in bridging the gap between accuracy and explainability in healthcare AI applications. By offering transparent and adaptable diagnostic models, our work contributes to the advancement of precision medicine and supports the development of equitable healthcare solutions. Future research will focus on extending these methods to larger and more diverse datasets to further validate their applicability across different medical conditions and populations.
Use of natural language processing to extract and classify papillary thyroid cancer features from surgical pathology reports
May 22, 2024



Abstract:Background We aim to use Natural Language Processing (NLP) to automate the extraction and classification of thyroid cancer risk factors from pathology reports. Methods We analyzed 1,410 surgical pathology reports from adult papillary thyroid cancer patients at Mayo Clinic, Rochester, MN, from 2010 to 2019. Structured and non-structured reports were used to create a consensus-based ground truth dictionary and categorized them into modified recurrence risk levels. Non-structured reports were narrative, while structured reports followed standardized formats. We then developed ThyroPath, a rule-based NLP pipeline, to extract and classify thyroid cancer features into risk categories. Training involved 225 reports (150 structured, 75 unstructured), with testing on 170 reports (120 structured, 50 unstructured) for evaluation. The pipeline's performance was assessed using both strict and lenient criteria for accuracy, precision, recall, and F1-score. Results In extraction tasks, ThyroPath achieved overall strict F-1 scores of 93% for structured reports and 90 for unstructured reports, covering 18 thyroid cancer pathology features. In classification tasks, ThyroPath-extracted information demonstrated an overall accuracy of 93% in categorizing reports based on their corresponding guideline-based risk of recurrence: 76.9% for high-risk, 86.8% for intermediate risk, and 100% for both low and very low-risk cases. However, ThyroPath achieved 100% accuracy across all thyroid cancer risk categories with human-extracted pathology information. Conclusions ThyroPath shows promise in automating the extraction and risk recurrence classification of thyroid pathology reports at large scale. It offers a solution to laborious manual reviews and advancing virtual registries. However, it requires further validation before implementation.
RxWhyQA: a clinical question-answering dataset with the challenge of multi-answer questions
Jan 07, 2022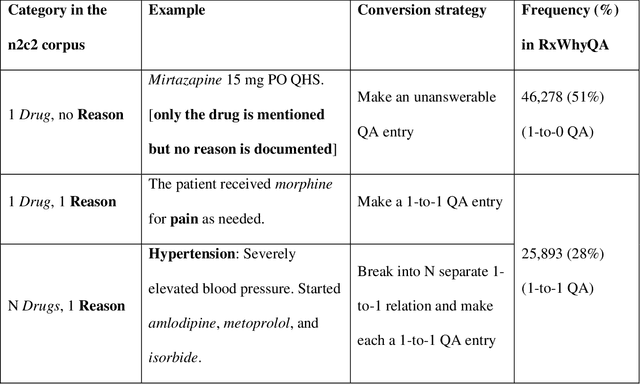
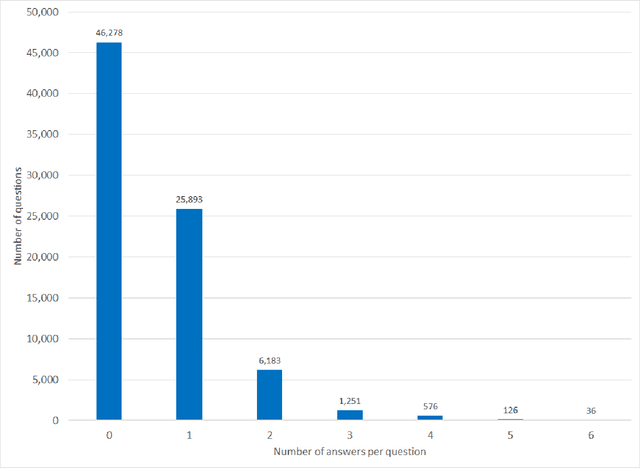


Abstract:Objectives Create a dataset for the development and evaluation of clinical question-answering (QA) systems that can handle multi-answer questions. Materials and Methods We leveraged the annotated relations from the 2018 National NLP Clinical Challenges (n2c2) corpus to generate a QA dataset. The 1-to-0 and 1-to-N drug-reason relations formed the unanswerable and multi-answer entries, which represent challenging scenarios lacking in the existing clinical QA datasets. Results The result RxWhyQA dataset contains 91,440 QA entries, of which half are unanswerable, and 21% (n=19,269) of the answerable ones require multiple answers. The dataset conforms to the community-vetted Stanford Question Answering Dataset (SQuAD) format. Discussion The RxWhyQA is useful for comparing different systems that need to handle the zero- and multi-answer challenges, demanding dual mitigation of both false positive and false negative answers. Conclusion We created and shared a clinical QA dataset with a focus on multi-answer questions to represent real-world scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge