Sungrim Moon
RxWhyQA: a clinical question-answering dataset with the challenge of multi-answer questions
Jan 07, 2022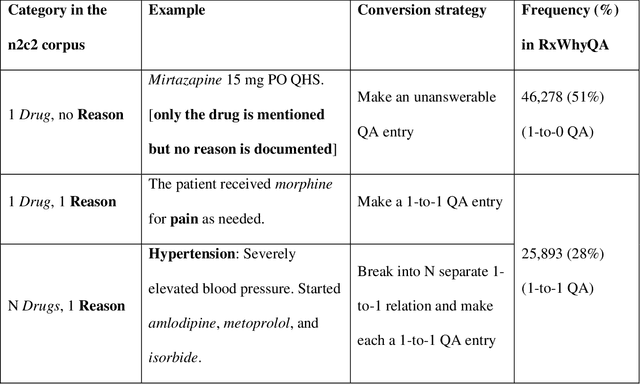
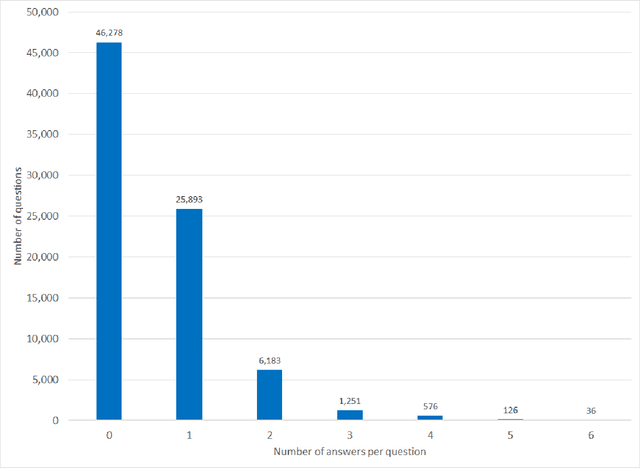


Abstract:Objectives Create a dataset for the development and evaluation of clinical question-answering (QA) systems that can handle multi-answer questions. Materials and Methods We leveraged the annotated relations from the 2018 National NLP Clinical Challenges (n2c2) corpus to generate a QA dataset. The 1-to-0 and 1-to-N drug-reason relations formed the unanswerable and multi-answer entries, which represent challenging scenarios lacking in the existing clinical QA datasets. Results The result RxWhyQA dataset contains 91,440 QA entries, of which half are unanswerable, and 21% (n=19,269) of the answerable ones require multiple answers. The dataset conforms to the community-vetted Stanford Question Answering Dataset (SQuAD) format. Discussion The RxWhyQA is useful for comparing different systems that need to handle the zero- and multi-answer challenges, demanding dual mitigation of both false positive and false negative answers. Conclusion We created and shared a clinical QA dataset with a focus on multi-answer questions to represent real-world scenarios.
Adapting and evaluating a deep learning language model for clinical why-question answering
Nov 13, 2019



Abstract:Objectives: To adapt and evaluate a deep learning language model for answering why-questions based on patient-specific clinical text. Materials and Methods: Bidirectional encoder representations from transformers (BERT) models were trained with varying data sources to perform SQuAD 2.0 style why-question answering (why-QA) on clinical notes. The evaluation focused on: 1) comparing the merits from different training data, 2) error analysis. Results: The best model achieved an accuracy of 0.707 (or 0.760 by partial match). Training toward customization for the clinical language helped increase 6% in accuracy. Discussion: The error analysis suggested that the model did not really perform deep reasoning and that clinical why-QA might warrant more sophisticated solutions. Conclusion: The BERT model achieved moderate accuracy in clinical why-QA and should benefit from the rapidly evolving technology. Despite the identified limitations, it could serve as a competent proxy for question-driven clinical information extraction.
Development of Clinical Concept Extraction Applications: A Methodology Review
Oct 28, 2019
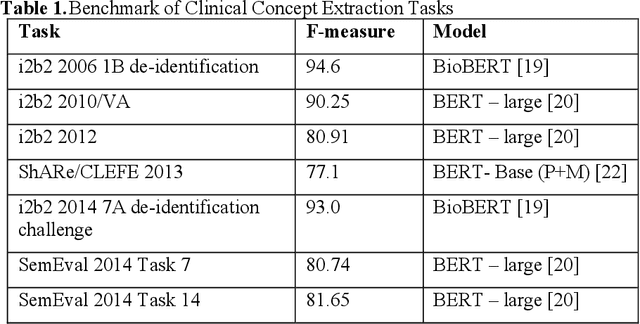
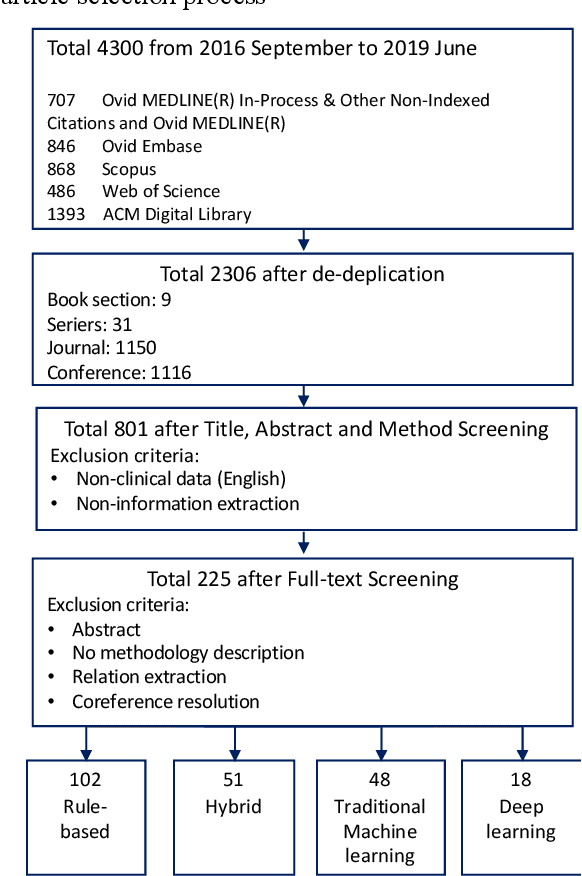
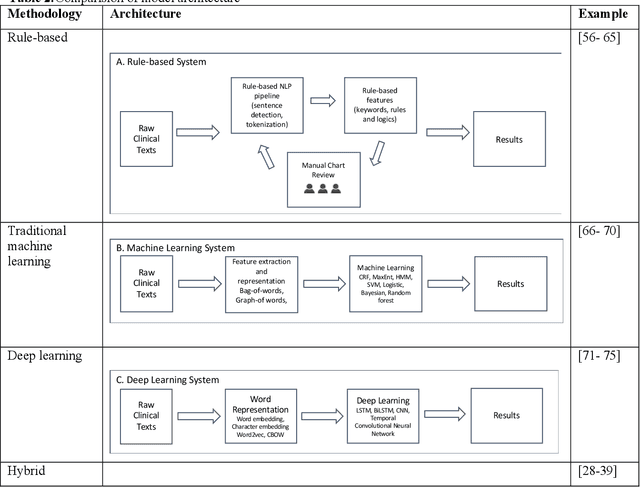
Abstract:Our study provided a review of the development of clinical concept extraction applications from January 2009 to June 2019. We hope, through the studying of different approaches with variant clinical context, can enhance the decision making for the development of clinical concept extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge