Yiqing Zhao
Unsupervised Learning to Subphenotype Delirium Patients from Electronic Health Records
Oct 31, 2021



Abstract:Delirium is a common acute onset brain dysfunction in the emergency setting and is associated with higher mortality. It is difficult to detect and monitor since its presentations and risk factors can be different depending on the underlying medical condition of patients. In our study, we aimed to identify subtypes within the delirium population and build subgroup-specific predictive models to detect delirium using Medical Information Mart for Intensive Care IV (MIMIC-IV) data. We showed that clusters exist within the delirium population. Differences in feature importance were also observed for subgroup-specific predictive models. Our work could recalibrate existing delirium prediction models for each delirium subgroup and improve the precision of delirium detection and monitoring for ICU or emergency department patients who had highly heterogeneous medical conditions.
Leveraging a Joint of Phenotypic and Genetic Features on Cancer Patient Subgrouping
Mar 30, 2021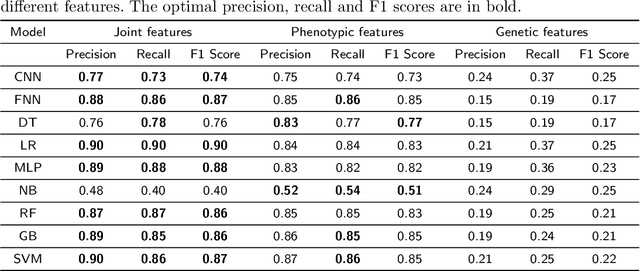
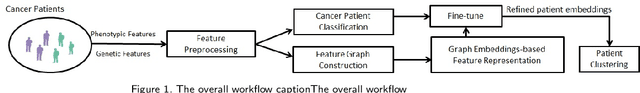
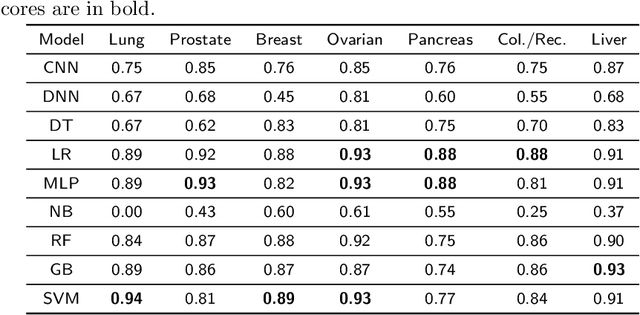
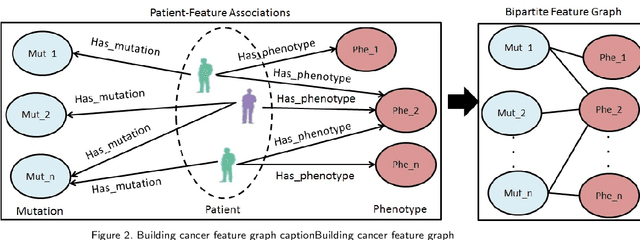
Abstract:Cancer is responsible for millions of deaths worldwide every year. Although significant progress has been achieved in cancer medicine, many issues remain to be addressed for improving cancer therapy. Appropriate cancer patient stratification is the prerequisite for selecting appropriate treatment plan, as cancer patients are of known heterogeneous genetic make-ups and phenotypic differences. In this study, built upon deep phenotypic characterizations extractable from Mayo Clinic electronic health records (EHRs) and genetic test reports for a collection of cancer patients, we developed a system leveraging a joint of phenotypic and genetic features for cancer patient subgrouping. The workflow is roughly divided into three parts: feature preprocessing, cancer patient classification, and cancer patient clustering based. In feature preprocessing step, we performed filtering, retaining the most relevant features. In cancer patient classification, we utilized joint categorical features to build a patient-feature matrix and applied nine different machine learning models, Random Forests (RF), Decision Tree (DT), Support Vector Machine (SVM), Naive Bayes (NB), Logistic Regression (LR), Multilayer Perceptron (MLP), Gradient Boosting (GB), Convolutional Neural Network (CNN), and Feedforward Neural Network (FNN), for classification purposes. Finally, in the cancer patient clustering step, we leveraged joint embeddings features and patient-feature associations to build an undirected feature graph and then trained the cancer feature node embeddings.
Comparisons of Graph Neural Networks on Cancer Classification Leveraging a Joint of Phenotypic and Genetic Features
Jan 14, 2021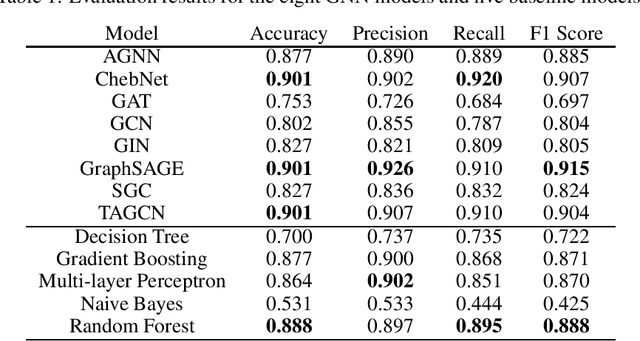
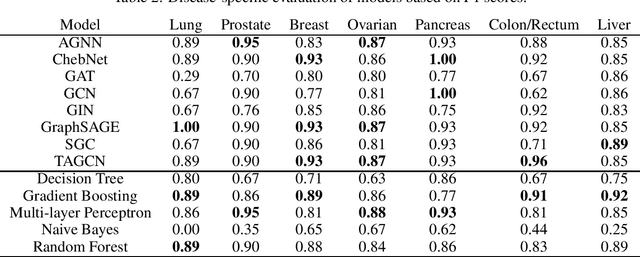
Abstract:Cancer is responsible for millions of deaths worldwide every year. Although significant progress hasbeen achieved in cancer medicine, many issues remain to be addressed for improving cancer therapy.Appropriate cancer patient stratification is the prerequisite for selecting appropriate treatment plan, ascancer patients are of known heterogeneous genetic make-ups and phenotypic differences. In thisstudy, built upon deep phenotypic characterizations extractable from Mayo Clinic electronic healthrecords (EHRs) and genetic test reports for a collection of cancer patients, we evaluated variousgraph neural networks (GNNs) leveraging a joint of phenotypic and genetic features for cancer typeclassification. Models were applied and fine-tuned on the Mayo Clinic cancer disease dataset. Theassessment was done through the reported accuracy, precision, recall, and F1 values as well as throughF1 scores based on the disease class. Per our evaluation results, GNNs on average outperformed thebaseline models with mean statistics always being higher that those of the baseline models (0.849 vs0.772 for accuracy, 0.858 vs 0.794 for precision, 0.843 vs 0.759 for recall, and 0.843 vs 0.855 for F1score). Among GNNs, ChebNet, GraphSAGE, and TAGCN showed the best performance, while GATshowed the worst. We applied and compared eight GNN models including AGNN, ChebNet, GAT,GCN, GIN, GraphSAGE, SGC, and TAGCN on the Mayo Clinic cancer disease dataset and assessedtheir performance as well as compared them with each other and with more conventional machinelearning models such as decision tree, gradient boosting, multi-layer perceptron, naive bayes, andrandom forest which we used as the baselines.
Development of Clinical Concept Extraction Applications: A Methodology Review
Oct 28, 2019
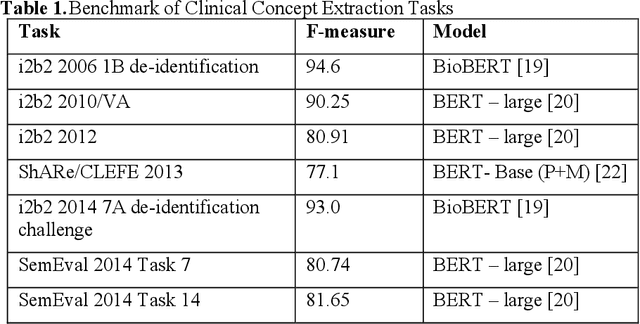
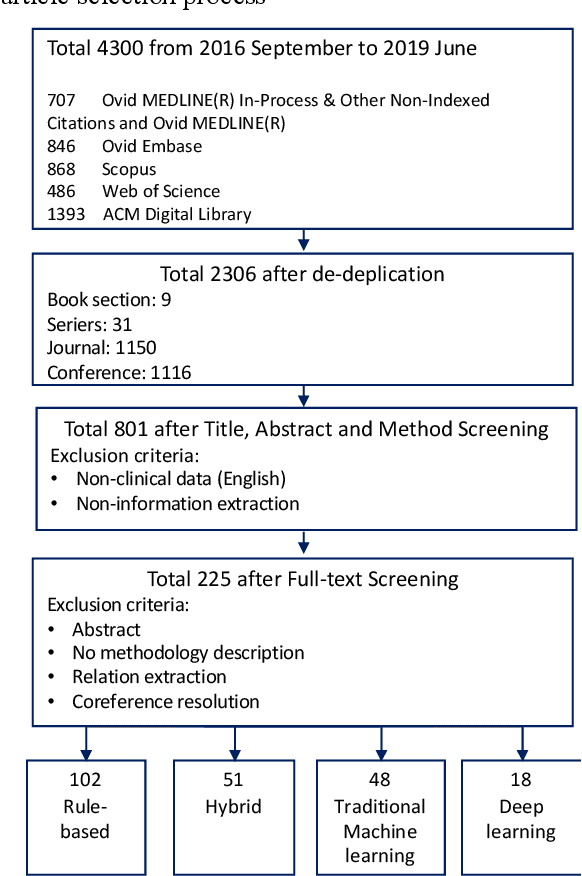
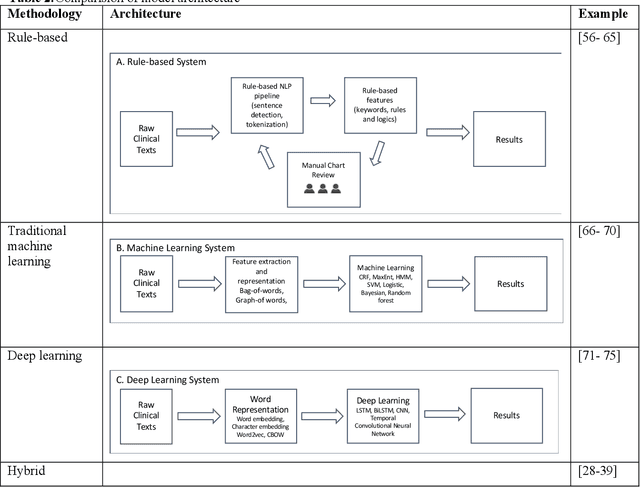
Abstract:Our study provided a review of the development of clinical concept extraction applications from January 2009 to June 2019. We hope, through the studying of different approaches with variant clinical context, can enhance the decision making for the development of clinical concept extraction.
Unsupervised Machine Learning for the Discovery of Latent Disease Clusters and Patient Subgroups Using Electronic Health Records
May 17, 2019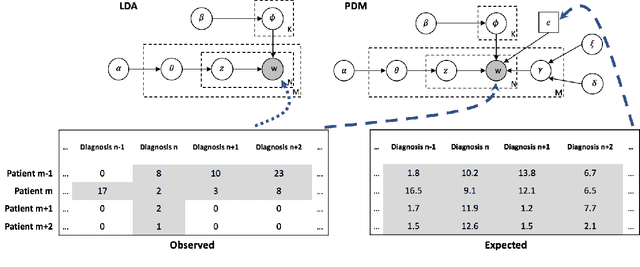
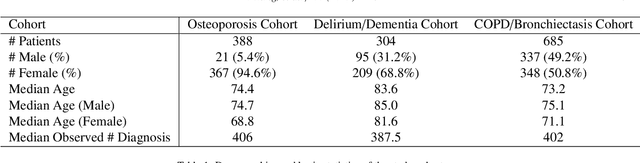
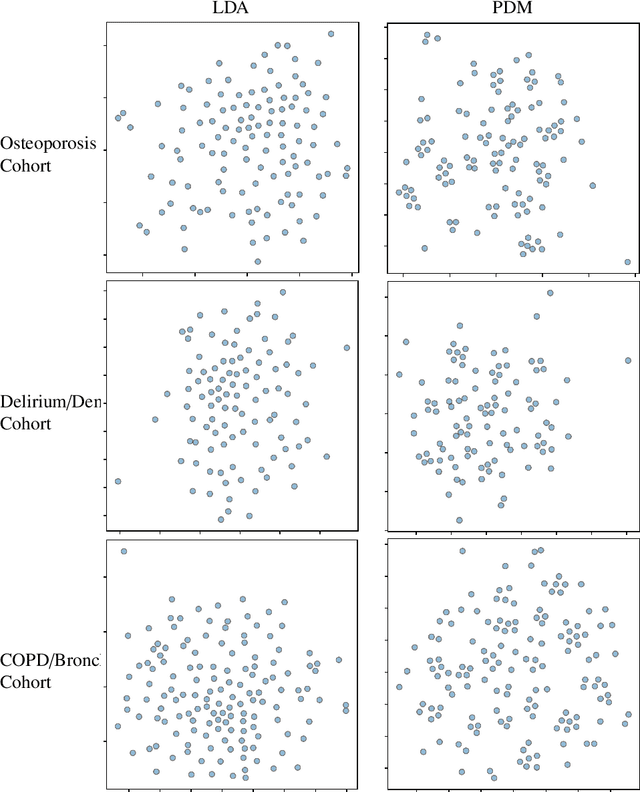
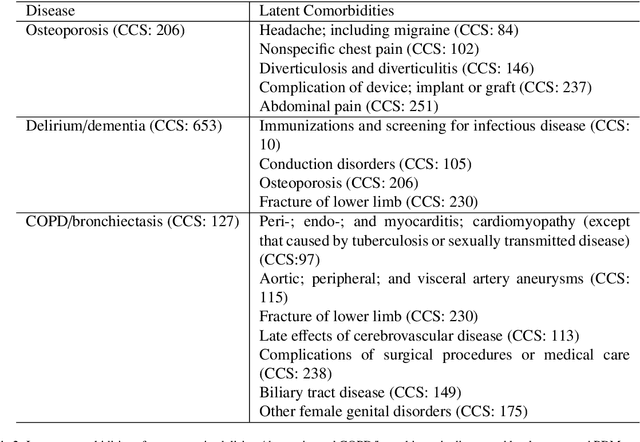
Abstract:Machine learning has become ubiquitous and a key technology on mining electronic health records (EHRs) for facilitating clinical research and practice. Unsupervised machine learning, as opposed to supervised learning, has shown promise in identifying novel patterns and relations from EHRs without using human created labels. In this paper, we investigate the application of unsupervised machine learning models in discovering latent disease clusters and patient subgroups based on EHRs. We utilized Latent Dirichlet Allocation (LDA), a generative probabilistic model, and proposed a novel model named Poisson Dirichlet Model (PDM), which extends the LDA approach using a Poisson distribution to model patients' disease diagnoses and to alleviate age and sex factors by considering both observed and expected observations. In the empirical experiments, we evaluated LDA and PDM on three patient cohorts with EHR data retrieved from the Rochester Epidemiology Project (REP), for the discovery of latent disease clusters and patient subgroups. We compared the effectiveness of LDA and PDM in identifying latent disease clusters through the visualization of disease representations learned by two approaches. We also tested the performance of LDA and PDM in differentiating patient subgroups through survival analysis, as well as statistical analysis. The experimental results show that the proposed PDM could effectively identify distinguished disease clusters by alleviating the impact of age and sex, and that LDA could stratify patients into more differentiable subgroups than PDM in terms of p-values. However, the subgroups discovered by PDM might imply the underlying patterns of diseases of greater interest in epidemiology research due to the alleviation of age and sex. Both unsupervised machine learning approaches could be leveraged to discover patient subgroups using EHRs but with different foci.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge