Ahmad P. Tafti
Explainable AI in Orthopedics: Challenges, Opportunities, and Prospects
Aug 09, 2023Abstract:While artificial intelligence (AI) has made many successful applications in various domains, its adoption in healthcare lags a little bit behind other high-stakes settings. Several factors contribute to this slower uptake, including regulatory frameworks, patient privacy concerns, and data heterogeneity. However, one significant challenge that impedes the implementation of AI in healthcare, particularly in orthopedics, is the lack of explainability and interpretability around AI models. Addressing the challenge of explainable AI (XAI) in orthopedics requires developing AI models and algorithms that prioritize transparency and interpretability, allowing clinicians, surgeons, and patients to understand the contributing factors behind any AI-powered predictive or descriptive models. The current contribution outlines several key challenges and opportunities that manifest in XAI in orthopedic practice. This work emphasizes the need for interdisciplinary collaborations between AI practitioners, orthopedic specialists, and regulatory entities to establish standards and guidelines for the adoption of XAI in orthopedics.
Learning Unbiased Image Segmentation: A Case Study with Plain Knee Radiographs
Aug 08, 2023Abstract:Automatic segmentation of knee bony anatomy is essential in orthopedics, and it has been around for several years in both pre-operative and post-operative settings. While deep learning algorithms have demonstrated exceptional performance in medical image analysis, the assessment of fairness and potential biases within these models remains limited. This study aims to revisit deep learning-powered knee-bony anatomy segmentation using plain radiographs to uncover visible gender and racial biases. The current contribution offers the potential to advance our understanding of biases, and it provides practical insights for researchers and practitioners in medical imaging. The proposed mitigation strategies mitigate gender and racial biases, ensuring fair and unbiased segmentation results. Furthermore, this work promotes equal access to accurate diagnoses and treatment outcomes for diverse patient populations, fostering equitable and inclusive healthcare provision.
Word Embedding Neural Networks to Advance Knee Osteoarthritis Research
Dec 22, 2022Abstract:Osteoarthritis (OA) is the most prevalent chronic joint disease worldwide, where knee OA takes more than 80% of commonly affected joints. Knee OA is not a curable disease yet, and it affects large columns of patients, making it costly to patients and healthcare systems. Etiology, diagnosis, and treatment of knee OA might be argued by variability in its clinical and physical manifestations. Although knee OA carries a list of well-known terminology aiming to standardize the nomenclature of the diagnosis, prognosis, treatment, and clinical outcomes of the chronic joint disease, in practice there is a wide range of terminology associated with knee OA across different data sources, including but not limited to biomedical literature, clinical notes, healthcare literacy, and health-related social media. Among these data sources, the scientific articles published in the biomedical literature usually make a principled pipeline to study disease. Rapid yet, accurate text mining on large-scale scientific literature may discover novel knowledge and terminology to better understand knee OA and to improve the quality of knee OA diagnosis, prevention, and treatment. The present works aim to utilize artificial neural network strategies to automatically extract vocabularies associated with knee OA diseases. Our finding indicates the feasibility of developing word embedding neural networks for autonomous keyword extraction and abstraction of knee OA.
Give me a knee radiograph, I will tell you where the knee joint area is: a deep convolutional neural network adventure
Feb 11, 2022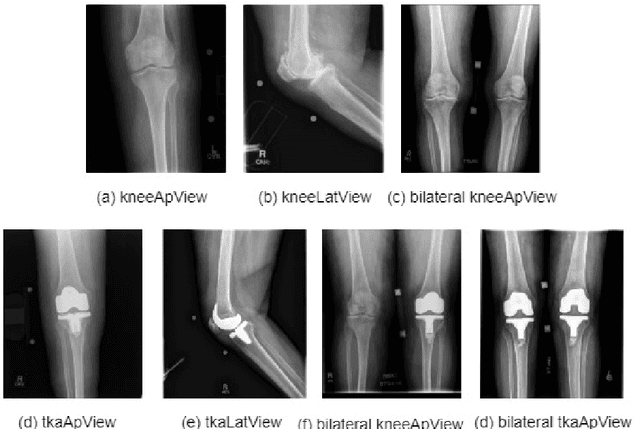

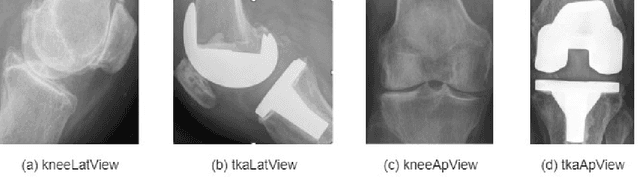
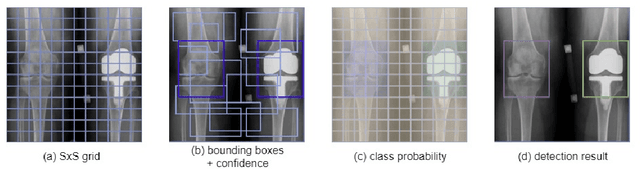
Abstract:Knee pain is undoubtedly the most common musculoskeletal symptom that impairs quality of life, confines mobility and functionality across all ages. Knee pain is clinically evaluated by routine radiographs, where the widespread adoption of radiographic images and their availability at low cost, make them the principle component in the assessment of knee pain and knee pathologies, such as arthritis, trauma, and sport injuries. However, interpretation of the knee radiographs is still highly subjective, and overlapping structures within the radiographs and the large volume of images needing to be analyzed on a daily basis, make interpretation challenging for both naive and experienced practitioners. There is thus a need to implement an artificial intelligence strategy to objectively and automatically interpret knee radiographs, facilitating triage of abnormal radiographs in a timely fashion. The current work proposes an accurate and effective pipeline for autonomous detection, localization, and classification of knee joint area in plain radiographs combining the You Only Look Once (YOLO v3) deep convolutional neural network with a large and fully-annotated knee radiographs dataset. The present work is expected to stimulate more interest from the deep learning computer vision community to this pragmatic and clinical application.
Unsupervised Machine Learning for the Discovery of Latent Disease Clusters and Patient Subgroups Using Electronic Health Records
May 17, 2019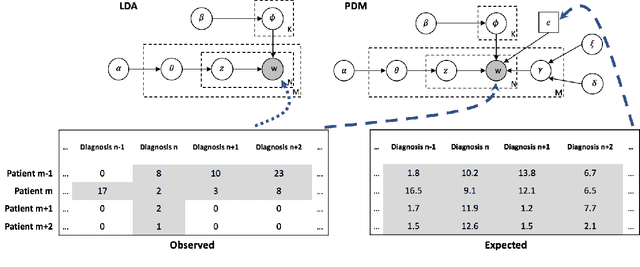
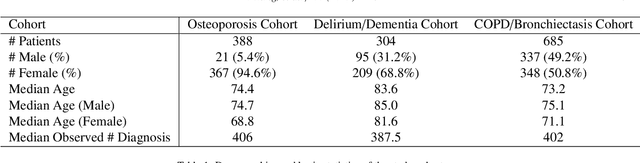
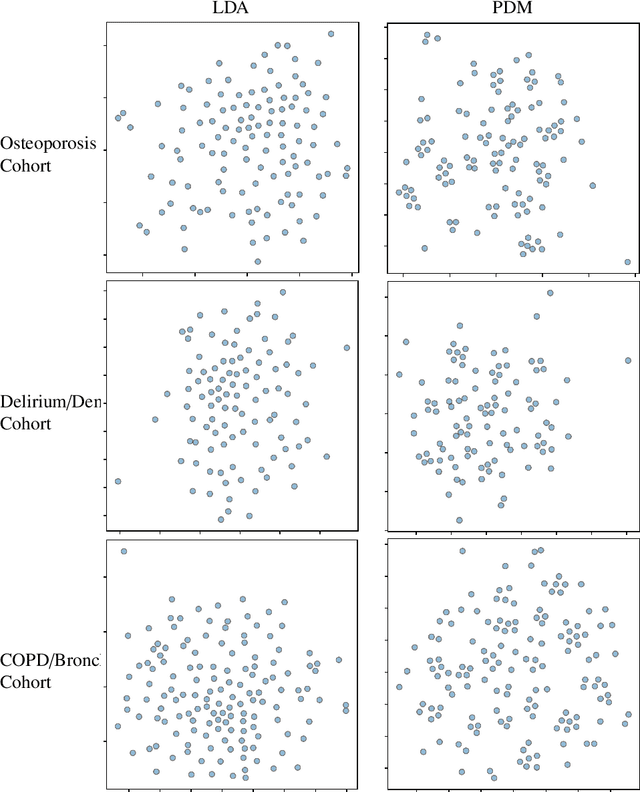
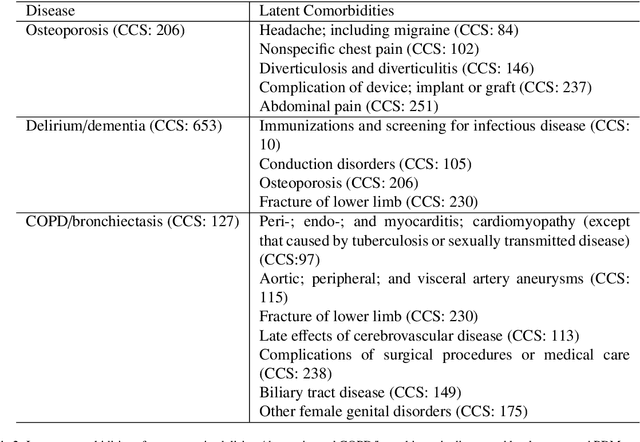
Abstract:Machine learning has become ubiquitous and a key technology on mining electronic health records (EHRs) for facilitating clinical research and practice. Unsupervised machine learning, as opposed to supervised learning, has shown promise in identifying novel patterns and relations from EHRs without using human created labels. In this paper, we investigate the application of unsupervised machine learning models in discovering latent disease clusters and patient subgroups based on EHRs. We utilized Latent Dirichlet Allocation (LDA), a generative probabilistic model, and proposed a novel model named Poisson Dirichlet Model (PDM), which extends the LDA approach using a Poisson distribution to model patients' disease diagnoses and to alleviate age and sex factors by considering both observed and expected observations. In the empirical experiments, we evaluated LDA and PDM on three patient cohorts with EHR data retrieved from the Rochester Epidemiology Project (REP), for the discovery of latent disease clusters and patient subgroups. We compared the effectiveness of LDA and PDM in identifying latent disease clusters through the visualization of disease representations learned by two approaches. We also tested the performance of LDA and PDM in differentiating patient subgroups through survival analysis, as well as statistical analysis. The experimental results show that the proposed PDM could effectively identify distinguished disease clusters by alleviating the impact of age and sex, and that LDA could stratify patients into more differentiable subgroups than PDM in terms of p-values. However, the subgroups discovered by PDM might imply the underlying patterns of diseases of greater interest in epidemiology research due to the alleviation of age and sex. Both unsupervised machine learning approaches could be leveraged to discover patient subgroups using EHRs but with different foci.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge