David Oniani
Ret.
Emerging Opportunities of Using Large Language Models for Translation Between Drug Molecules and Indications
Feb 16, 2024Abstract:A drug molecule is a substance that changes the organism's mental or physical state. Every approved drug has an indication, which refers to the therapeutic use of that drug for treating a particular medical condition. While the Large Language Model (LLM), a generative Artificial Intelligence (AI) technique, has recently demonstrated effectiveness in translating between molecules and their textual descriptions, there remains a gap in research regarding their application in facilitating the translation between drug molecules and indications, or vice versa, which could greatly benefit the drug discovery process. The capability of generating a drug from a given indication would allow for the discovery of drugs targeting specific diseases or targets and ultimately provide patients with better treatments. In this paper, we first propose a new task, which is the translation between drug molecules and corresponding indications, and then test existing LLMs on this new task. Specifically, we consider nine variations of the T5 LLM and evaluate them on two public datasets obtained from ChEMBL and DrugBank. Our experiments show the early results of using LLMs for this task and provide a perspective on the state-of-the-art. We also emphasize the current limitations and discuss future work that has the potential to improve the performance on this task. The creation of molecules from indications, or vice versa, will allow for more efficient targeting of diseases and significantly reduce the cost of drug discovery, with the potential to revolutionize the field of drug discovery in the era of generative AI.
Enhancing Large Language Models for Clinical Decision Support by Incorporating Clinical Practice Guidelines
Jan 23, 2024



Abstract:Background Large Language Models (LLMs), enhanced with Clinical Practice Guidelines (CPGs), can significantly improve Clinical Decision Support (CDS). However, methods for incorporating CPGs into LLMs are not well studied. Methods We develop three distinct methods for incorporating CPGs into LLMs: Binary Decision Tree (BDT), Program-Aided Graph Construction (PAGC), and Chain-of-Thought-Few-Shot Prompting (CoT-FSP). To evaluate the effectiveness of the proposed methods, we create a set of synthetic patient descriptions and conduct both automatic and human evaluation of the responses generated by four LLMs: GPT-4, GPT-3.5 Turbo, LLaMA, and PaLM 2. Zero-Shot Prompting (ZSP) was used as the baseline method. We focus on CDS for COVID-19 outpatient treatment as the case study. Results All four LLMs exhibit improved performance when enhanced with CPGs compared to the baseline ZSP. BDT outperformed both CoT-FSP and PAGC in automatic evaluation. All of the proposed methods demonstrated high performance in human evaluation. Conclusion LLMs enhanced with CPGs demonstrate superior performance, as compared to plain LLMs with ZSP, in providing accurate recommendations for COVID-19 outpatient treatment, which also highlights the potential for broader applications beyond the case study.
In-Context Learning Functions with Varying Number of Minima
Nov 22, 2023Abstract:Large Language Models (LLMs) have proven effective at In-Context Learning (ICL), an ability that allows them to create predictors from labeled examples. Few studies have explored the interplay between ICL and specific properties of functions it attempts to approximate. In our study, we use a formal framework to explore ICL and propose a new task of approximating functions with varying number of minima. We implement a method that allows for producing functions with given inputs as minima. We find that increasing the number of minima degrades ICL performance. At the same time, our evaluation shows that ICL outperforms 2-layer Neural Network (2NN) model. Furthermore, ICL learns faster than 2NN in all settings. We validate the findings through a set of few-shot experiments across various hyperparameter configurations.
Foundation Metrics: Quantifying Effectiveness of Healthcare Conversations powered by Generative AI
Sep 21, 2023Abstract:Generative Artificial Intelligence is set to revolutionize healthcare delivery by transforming traditional patient care into a more personalized, efficient, and proactive process. Chatbots, serving as interactive conversational models, will probably drive this patient-centered transformation in healthcare. Through the provision of various services, including diagnosis, personalized lifestyle recommendations, and mental health support, the objective is to substantially augment patient health outcomes, all the while mitigating the workload burden on healthcare providers. The life-critical nature of healthcare applications necessitates establishing a unified and comprehensive set of evaluation metrics for conversational models. Existing evaluation metrics proposed for various generic large language models (LLMs) demonstrate a lack of comprehension regarding medical and health concepts and their significance in promoting patients' well-being. Moreover, these metrics neglect pivotal user-centered aspects, including trust-building, ethics, personalization, empathy, user comprehension, and emotional support. The purpose of this paper is to explore state-of-the-art LLM-based evaluation metrics that are specifically applicable to the assessment of interactive conversational models in healthcare. Subsequently, we present an comprehensive set of evaluation metrics designed to thoroughly assess the performance of healthcare chatbots from an end-user perspective. These metrics encompass an evaluation of language processing abilities, impact on real-world clinical tasks, and effectiveness in user-interactive conversations. Finally, we engage in a discussion concerning the challenges associated with defining and implementing these metrics, with particular emphasis on confounding factors such as the target audience, evaluation methods, and prompt techniques involved in the evaluation process.
Large Language Models Vote: Prompting for Rare Disease Identification
Aug 28, 2023



Abstract:The emergence of generative Large Language Models (LLMs) emphasizes the need for accurate and efficient prompting approaches. LLMs are often applied in Few-Shot Learning (FSL) contexts, where tasks are executed with minimal training data. FSL has become popular in many Artificial Intelligence (AI) subdomains, including AI for health. Rare diseases affect a small fraction of the population. Rare disease identification from clinical notes inherently requires FSL techniques due to limited data availability. Manual data collection and annotation is both expensive and time-consuming. In this paper, we propose Models-Vote Prompting (MVP), a flexible prompting approach for improving the performance of LLM queries in FSL settings. MVP works by prompting numerous LLMs to perform the same tasks and then conducting a majority vote on the resulting outputs. This method achieves improved results to any one model in the ensemble on one-shot rare disease identification and classification tasks. We also release a novel rare disease dataset for FSL, available to those who signed the MIMIC-IV Data Use Agreement (DUA). Furthermore, in using MVP, each model is prompted multiple times, substantially increasing the time needed for manual annotation, and to address this, we assess the feasibility of using JSON for automating generative LLM evaluation.
From Military to Healthcare: Adopting and Expanding Ethical Principles for Generative Artificial Intelligence
Aug 04, 2023Abstract:In 2020, the U.S. Department of Defense officially disclosed a set of ethical principles to guide the use of Artificial Intelligence (AI) technologies on future battlefields. Despite stark differences, there are core similarities between the military and medical service. Warriors on battlefields often face life-altering circumstances that require quick decision-making. Medical providers experience similar challenges in a rapidly changing healthcare environment, such as in the emergency department or during surgery treating a life-threatening condition. Generative AI, an emerging technology designed to efficiently generate valuable information, holds great promise. As computing power becomes more accessible and the abundance of health data, such as electronic health records, electrocardiograms, and medical images, increases, it is inevitable that healthcare will be revolutionized by this technology. Recently, generative AI has captivated the research community, leading to debates about its application in healthcare, mainly due to concerns about transparency and related issues. Meanwhile, concerns about the potential exacerbation of health disparities due to modeling biases have raised notable ethical concerns regarding the use of this technology in healthcare. However, the ethical principles for generative AI in healthcare have been understudied, and decision-makers often fail to consider the significance of generative AI. In this paper, we propose GREAT PLEA ethical principles, encompassing governance, reliability, equity, accountability, traceability, privacy, lawfulness, empathy, and autonomy, for generative AI in healthcare. We aim to proactively address the ethical dilemmas and challenges posed by the integration of generative AI in healthcare.
ReDWINE: A Clinical Datamart with Text Analytical Capabilities to Facilitate Rehabilitation Research
Apr 12, 2023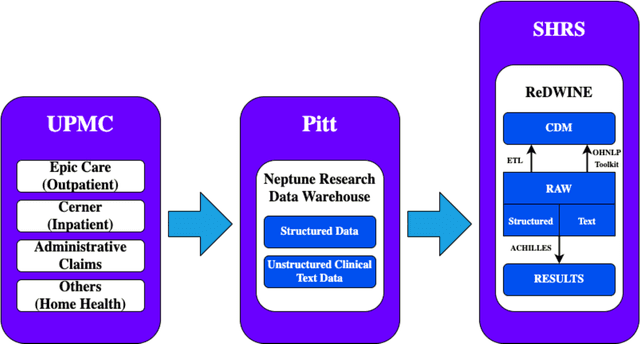
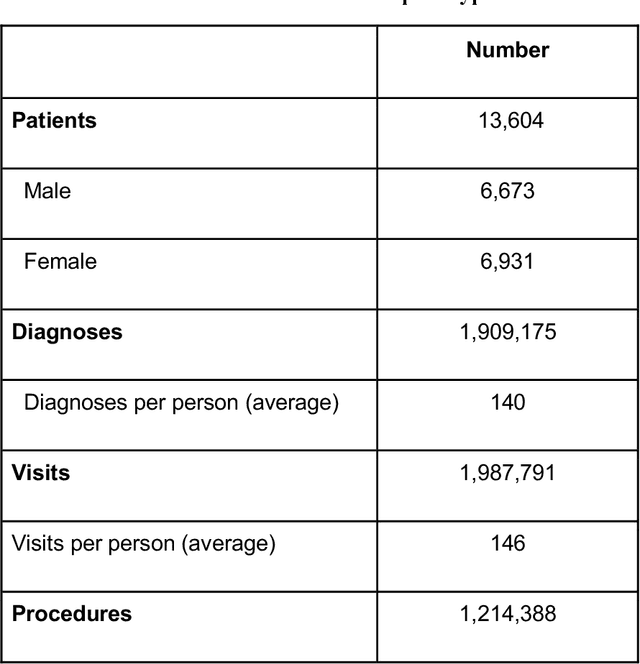
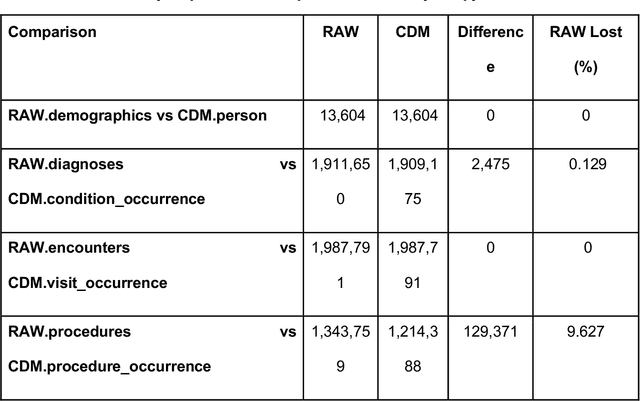
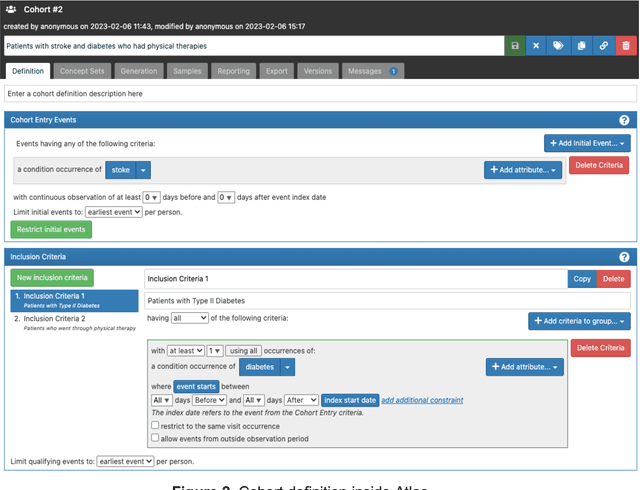
Abstract:Rehabilitation research focuses on determining the components of a treatment intervention, the mechanism of how these components lead to recovery and rehabilitation, and ultimately the optimal intervention strategies to maximize patients' physical, psychologic, and social functioning. Traditional randomized clinical trials that study and establish new interventions face several challenges, such as high cost and time commitment. Observational studies that use existing clinical data to observe the effect of an intervention have shown several advantages over RCTs. Electronic Health Records (EHRs) have become an increasingly important resource for conducting observational studies. To support these studies, we developed a clinical research datamart, called ReDWINE (Rehabilitation Datamart With Informatics iNfrastructure for rEsearch), that transforms the rehabilitation-related EHR data collected from the UPMC health care system to the Observational Health Data Sciences and Informatics (OHDSI) Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) to facilitate rehabilitation research. The standardized EHR data stored in ReDWINE will further reduce the time and effort required by investigators to pool, harmonize, clean, and analyze data from multiple sources, leading to more robust and comprehensive research findings. ReDWINE also includes deployment of data visualization and data analytics tools to facilitate cohort definition and clinical data analysis. These include among others the Open Health Natural Language Processing (OHNLP) toolkit, a high-throughput NLP pipeline, to provide text analytical capabilities at scale in ReDWINE. Using this comprehensive representation of patient data in ReDWINE for rehabilitation research will facilitate real-world evidence for health interventions and outcomes.
Toward Improving Health Literacy in Patient Education Materials with Neural Machine Translation Models
Sep 14, 2022
Abstract:Health literacy is the central focus of Healthy People 2030, the fifth iteration of the U.S. national goals and objectives. People with low health literacy usually have trouble understanding health information, following post-visit instructions, and using prescriptions, which results in worse health outcomes and serious health disparities. In this study, we propose to leverage natural language processing techniques to improve health literacy in patient education materials by automatically translating illiterate languages in a given sentence. We scraped patient education materials from four online health information websites: MedlinePlus.gov, Drugs.com, Mayoclinic.org and Reddit.com. We trained and tested the state-of-the-art neural machine translation (NMT) models on a silver standard training dataset and a gold standard testing dataset, respectively. The experimental results showed that the Bidirectional Long Short-Term Memory (BiLSTM) NMT model outperformed Bidirectional Encoder Representations from Transformers (BERT)-based NMT models. We also verified the effectiveness of NMT models in translating health illiterate languages by comparing the ratio of health illiterate language in the sentence. The proposed NMT models were able to identify the correct complicated words and simplify into layman language while at the same time the models suffer from sentence completeness, fluency, readability, and have difficulty in translating certain medical terms.
Few-Shot Learning for Clinical Natural Language Processing Using Siamese Neural Networks
Aug 31, 2022



Abstract:Clinical Natural Language Processing (NLP) has become an emerging technology in healthcare that leverages a large amount of free-text data in electronic health records (EHRs) to improve patient care, support clinical decisions, and facilitate clinical and translational science research. Deep learning has achieved state-of-the-art performance in many clinical NLP tasks. However, training deep learning models usually require large annotated datasets, which are normally not publicly available and can be time-consuming to build in clinical domains. Working with smaller annotated datasets is typical in clinical NLP and therefore, ensuring that deep learning models perform well is crucial for the models to be used in real-world applications. A widely adopted approach is fine-tuning existing Pre-trained Language Models (PLMs), but these attempts fall short when the training dataset contains only a few annotated samples. Few-Shot Learning (FSL) has recently been investigated to tackle this problem. Siamese Neural Network (SNN) has been widely utilized as an FSL approach in computer vision, but has not been studied well in NLP. Furthermore, the literature on its applications in clinical domains is scarce. In this paper, we propose two SNN-based FSL approaches for clinical NLP, including pre-trained SNN (PT-SNN) and SNN with second-order embeddings (SOE-SNN). We evaluated the proposed approaches on two clinical tasks, namely clinical text classification and clinical named entity recognition. We tested three few-shot settings including 4-shot, 8-shot, and 16-shot learning. Both clinical NLP tasks were benchmarked using three PLMs, including BERT, BioBERT, and BioClinicalBERT. The experimental results verified the effectiveness of the proposed SNN-based FSL approaches in both clinical NLP tasks.
Leveraging a Joint of Phenotypic and Genetic Features on Cancer Patient Subgrouping
Mar 30, 2021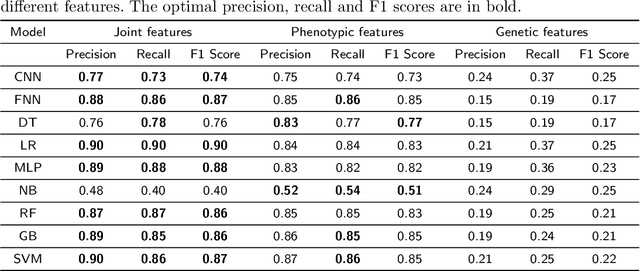
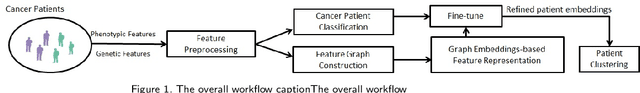
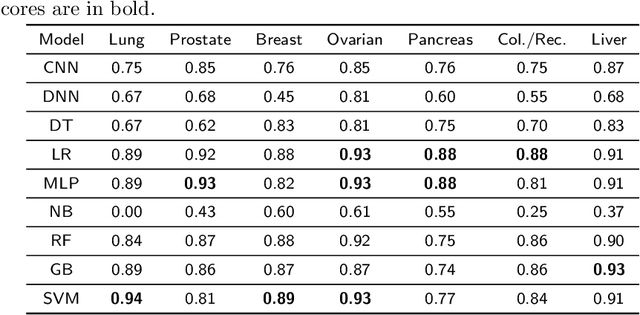
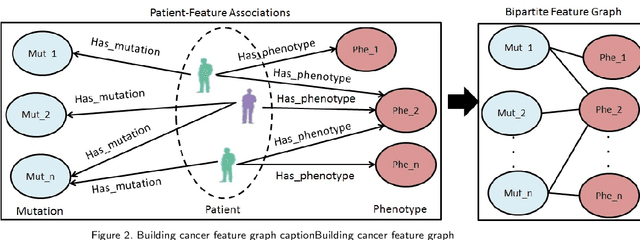
Abstract:Cancer is responsible for millions of deaths worldwide every year. Although significant progress has been achieved in cancer medicine, many issues remain to be addressed for improving cancer therapy. Appropriate cancer patient stratification is the prerequisite for selecting appropriate treatment plan, as cancer patients are of known heterogeneous genetic make-ups and phenotypic differences. In this study, built upon deep phenotypic characterizations extractable from Mayo Clinic electronic health records (EHRs) and genetic test reports for a collection of cancer patients, we developed a system leveraging a joint of phenotypic and genetic features for cancer patient subgrouping. The workflow is roughly divided into three parts: feature preprocessing, cancer patient classification, and cancer patient clustering based. In feature preprocessing step, we performed filtering, retaining the most relevant features. In cancer patient classification, we utilized joint categorical features to build a patient-feature matrix and applied nine different machine learning models, Random Forests (RF), Decision Tree (DT), Support Vector Machine (SVM), Naive Bayes (NB), Logistic Regression (LR), Multilayer Perceptron (MLP), Gradient Boosting (GB), Convolutional Neural Network (CNN), and Feedforward Neural Network (FNN), for classification purposes. Finally, in the cancer patient clustering step, we leveraged joint embeddings features and patient-feature associations to build an undirected feature graph and then trained the cancer feature node embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge