Olivier Gevaert
SAGE-FM: A lightweight and interpretable spatial transcriptomics foundation model
Jan 21, 2026Abstract:Spatial transcriptomics enables spatial gene expression profiling, motivating computational models that capture spatially conditioned regulatory relationships. We introduce SAGE-FM, a lightweight spatial transcriptomics foundation model based on graph convolutional networks (GCNs) trained with a masked central spot prediction objective. Trained on 416 human Visium samples spanning 15 organs, SAGE-FM learns spatially coherent embeddings that robustly recover masked genes, with 91% of masked genes showing significant correlations (p < 0.05). The embeddings generated by SAGE-FM outperform MOFA and existing spatial transcriptomics methods in unsupervised clustering and preservation of biological heterogeneity. SAGE-FM generalizes to downstream tasks, enabling 81% accuracy in pathologist-defined spot annotation in oropharyngeal squamous cell carcinoma and improving glioblastoma subtype prediction relative to MOFA. In silico perturbation experiments further demonstrate that the model captures directional ligand-receptor and upstream-downstream regulatory effects consistent with ground truth. These results demonstrate that simple, parameter-efficient GCNs can serve as biologically interpretable and spatially aware foundation models for large-scale spatial transcriptomics.
Benchmarking Chest X-ray Diagnosis Models Across Multinational Datasets
May 21, 2025Abstract:Foundation models leveraging vision-language pretraining have shown promise in chest X-ray (CXR) interpretation, yet their real-world performance across diverse populations and diagnostic tasks remains insufficiently evaluated. This study benchmarks the diagnostic performance and generalizability of foundation models versus traditional convolutional neural networks (CNNs) on multinational CXR datasets. We evaluated eight CXR diagnostic models - five vision-language foundation models and three CNN-based architectures - across 37 standardized classification tasks using six public datasets from the USA, Spain, India, and Vietnam, and three private datasets from hospitals in China. Performance was assessed using AUROC, AUPRC, and other metrics across both shared and dataset-specific tasks. Foundation models outperformed CNNs in both accuracy and task coverage. MAVL, a model incorporating knowledge-enhanced prompts and structured supervision, achieved the highest performance on public (mean AUROC: 0.82; AUPRC: 0.32) and private (mean AUROC: 0.95; AUPRC: 0.89) datasets, ranking first in 14 of 37 public and 3 of 4 private tasks. All models showed reduced performance on pediatric cases, with average AUROC dropping from 0.88 +/- 0.18 in adults to 0.57 +/- 0.29 in children (p = 0.0202). These findings highlight the value of structured supervision and prompt design in radiologic AI and suggest future directions including geographic expansion and ensemble modeling for clinical deployment. Code for all evaluated models is available at https://drive.google.com/drive/folders/1B99yMQm7bB4h1sVMIBja0RfUu8gLktCE
Prior Knowledge Injection into Deep Learning Models Predicting Gene Expression from Whole Slide Images
Jan 23, 2025Abstract:Cancer diagnosis and prognosis primarily depend on clinical parameters such as age and tumor grade, and are increasingly complemented by molecular data, such as gene expression, from tumor sequencing. However, sequencing is costly and delays oncology workflows. Recent advances in Deep Learning allow to predict molecular information from morphological features within Whole Slide Images (WSIs), offering a cost-effective proxy of the molecular markers. While promising, current methods lack the robustness to fully replace direct sequencing. Here we aim to improve existing methods by introducing a model-agnostic framework that allows to inject prior knowledge on gene-gene interactions into Deep Learning architectures, thereby increasing accuracy and robustness. We design the framework to be generic and flexibly adaptable to a wide range of architectures. In a case study on breast cancer, our strategy leads to an average increase of 983 significant genes (out of 25,761) across all 18 experiments, with 14 generalizing to an increase on an independent dataset. Our findings reveal a high potential for injection of prior knowledge to increase gene expression prediction performance from WSIs across a wide range of architectures.
Identification of head impact locations, speeds, and force based on head kinematics
Sep 12, 2024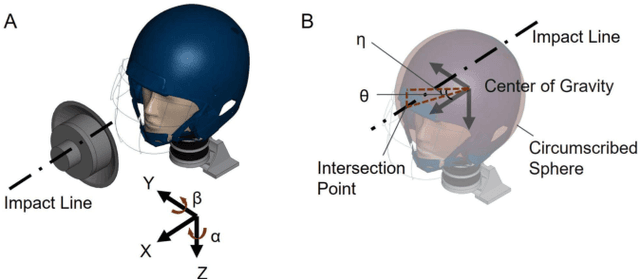
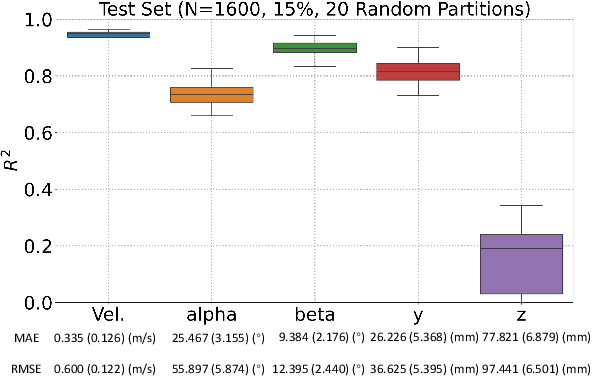
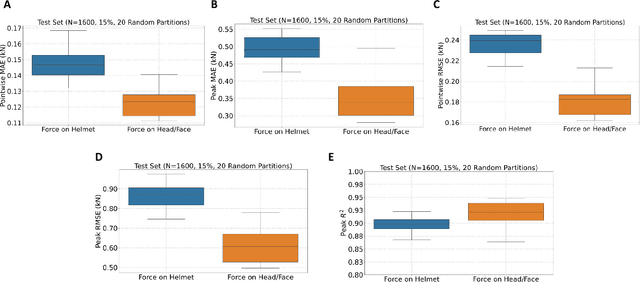
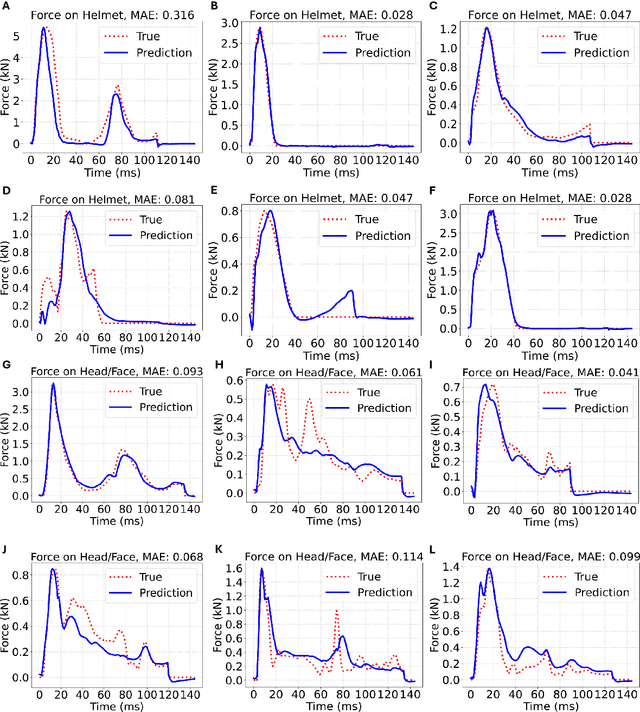
Abstract:Objective: Head impact information including impact directions, speeds and force are important to study traumatic brain injury, design and evaluate protective gears. This study presents a deep learning model developed to accurately predict head impact information, including location, speed, orientation, and force, based on head kinematics during helmeted impacts. Methods: Leveraging a dataset of 16,000 simulated helmeted head impacts using the Riddell helmet finite element model, we implemented a Long Short-Term Memory (LSTM) network to process the head kinematics: tri-axial linear accelerations and angular velocities. Results: The models accurately predict the impact parameters describing impact location, direction, speed, and the impact force profile with R2 exceeding 70% for all tasks. Further validation was conducted using an on-field dataset recorded by instrumented mouthguards and videos, consisting of 79 head impacts in which the impact location can be clearly identified. The deep learning model significantly outperformed existing methods, achieving a 79.7% accuracy in identifying impact locations, compared to lower accuracies with traditional methods (the highest accuracy of existing methods is 49.4%). Conclusion: The precision underscores the model's potential in enhancing helmet design and safety in sports by providing more accurate impact data. Future studies should test the models across various helmets and sports on large in vivo datasets to validate the accuracy of the models, employing techniques like transfer learning to broaden its effectiveness.
Unraveling Radiomics Complexity: Strategies for Optimal Simplicity in Predictive Modeling
Jul 05, 2024



Abstract:Background: The high dimensionality of radiomic feature sets, the variability in radiomic feature types and potentially high computational requirements all underscore the need for an effective method to identify the smallest set of predictive features for a given clinical problem. Purpose: Develop a methodology and tools to identify and explain the smallest set of predictive radiomic features. Materials and Methods: 89,714 radiomic features were extracted from five cancer datasets: low-grade glioma, meningioma, non-small cell lung cancer (NSCLC), and two renal cell carcinoma cohorts (n=2104). Features were categorized by computational complexity into morphological, intensity, texture, linear filters, and nonlinear filters. Models were trained and evaluated on each complexity level using the area under the curve (AUC). The most informative features were identified, and their importance was explained. The optimal complexity level and associated most informative features were identified using systematic statistical significance analyses and a false discovery avoidance procedure, respectively. Their predictive importance was explained using a novel tree-based method. Results: MEDimage, a new open-source tool, was developed to facilitate radiomic studies. Morphological features were optimal for MRI-based meningioma (AUC: 0.65) and low-grade glioma (AUC: 0.68). Intensity features were optimal for CECT-based renal cell carcinoma (AUC: 0.82) and CT-based NSCLC (AUC: 0.76). Texture features were optimal for MRI-based renal cell carcinoma (AUC: 0.72). Tuning the Hounsfield unit range improved results for CECT-based renal cell carcinoma (AUC: 0.86). Conclusion: Our proposed methodology and software can estimate the optimal radiomics complexity level for specific medical outcomes, potentially simplifying the use of radiomics in predictive modeling across various contexts.
Towards a more inductive world for drug repurposing approaches
Nov 24, 2023Abstract:Drug-target interaction (DTI) prediction is a challenging, albeit essential task in drug repurposing. Learning on graph models have drawn special attention as they can significantly reduce drug repurposing costs and time commitment. However, many current approaches require high-demanding additional information besides DTIs that complicates their evaluation process and usability. Additionally, structural differences in the learning architecture of current models hinder their fair benchmarking. In this work, we first perform an in-depth evaluation of current DTI datasets and prediction models through a robust benchmarking process, and show that DTI prediction methods based on transductive models lack generalization and lead to inflated performance when evaluated as previously done in the literature, hence not being suited for drug repurposing approaches. We then propose a novel biologically-driven strategy for negative edge subsampling and show through in vitro validation that newly discovered interactions are indeed true. We envision this work as the underpinning for future fair benchmarking and robust model design. All generated resources and tools are publicly available as a python package.
Multimodal Machine Learning in Image-Based and Clinical Biomedicine: Survey and Prospects
Nov 20, 2023Abstract:Machine learning (ML) applications in medical artificial intelligence (AI) systems have shifted from traditional and statistical methods to increasing application of deep learning models. This survey navigates the current landscape of multimodal ML, focusing on its profound impact on medical image analysis and clinical decision support systems. Emphasizing challenges and innovations in addressing multimodal representation, fusion, translation, alignment, and co-learning, the paper explores the transformative potential of multimodal models for clinical predictions. It also questions practical implementation of such models, bringing attention to the dynamics between decision support systems and healthcare providers. Despite advancements, challenges such as data biases and the scarcity of "big data" in many biomedical domains persist. We conclude with a discussion on effective innovation and collaborative efforts to further the miss
Foundation Metrics: Quantifying Effectiveness of Healthcare Conversations powered by Generative AI
Sep 21, 2023Abstract:Generative Artificial Intelligence is set to revolutionize healthcare delivery by transforming traditional patient care into a more personalized, efficient, and proactive process. Chatbots, serving as interactive conversational models, will probably drive this patient-centered transformation in healthcare. Through the provision of various services, including diagnosis, personalized lifestyle recommendations, and mental health support, the objective is to substantially augment patient health outcomes, all the while mitigating the workload burden on healthcare providers. The life-critical nature of healthcare applications necessitates establishing a unified and comprehensive set of evaluation metrics for conversational models. Existing evaluation metrics proposed for various generic large language models (LLMs) demonstrate a lack of comprehension regarding medical and health concepts and their significance in promoting patients' well-being. Moreover, these metrics neglect pivotal user-centered aspects, including trust-building, ethics, personalization, empathy, user comprehension, and emotional support. The purpose of this paper is to explore state-of-the-art LLM-based evaluation metrics that are specifically applicable to the assessment of interactive conversational models in healthcare. Subsequently, we present an comprehensive set of evaluation metrics designed to thoroughly assess the performance of healthcare chatbots from an end-user perspective. These metrics encompass an evaluation of language processing abilities, impact on real-world clinical tasks, and effectiveness in user-interactive conversations. Finally, we engage in a discussion concerning the challenges associated with defining and implementing these metrics, with particular emphasis on confounding factors such as the target audience, evaluation methods, and prompt techniques involved in the evaluation process.
Reliability-based cleaning of noisy training labels with inductive conformal prediction in multi-modal biomedical data mining
Sep 13, 2023Abstract:Accurately labeling biomedical data presents a challenge. Traditional semi-supervised learning methods often under-utilize available unlabeled data. To address this, we propose a novel reliability-based training data cleaning method employing inductive conformal prediction (ICP). This method capitalizes on a small set of accurately labeled training data and leverages ICP-calculated reliability metrics to rectify mislabeled data and outliers within vast quantities of noisy training data. The efficacy of the method is validated across three classification tasks within distinct modalities: filtering drug-induced-liver-injury (DILI) literature with title and abstract, predicting ICU admission of COVID-19 patients through CT radiomics and electronic health records, and subtyping breast cancer using RNA-sequencing data. Varying levels of noise to the training labels were introduced through label permutation. Results show significant enhancements in classification performance: accuracy enhancement in 86 out of 96 DILI experiments (up to 11.4%), AUROC and AUPRC enhancements in all 48 COVID-19 experiments (up to 23.8% and 69.8%), and accuracy and macro-average F1 score improvements in 47 out of 48 RNA-sequencing experiments (up to 74.6% and 89.0%). Our method offers the potential to substantially boost classification performance in multi-modal biomedical machine learning tasks. Importantly, it accomplishes this without necessitating an excessive volume of meticulously curated training data.
Toward more accurate and generalizable brain deformation estimators for traumatic brain injury detection with unsupervised domain adaptation
Jun 08, 2023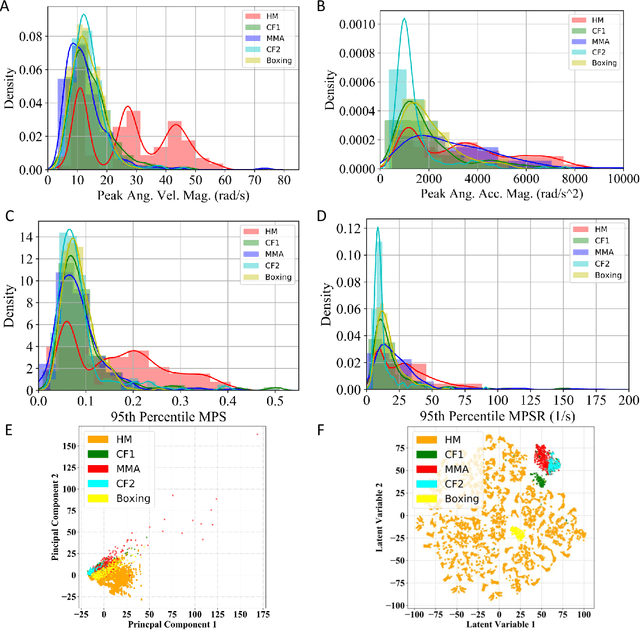
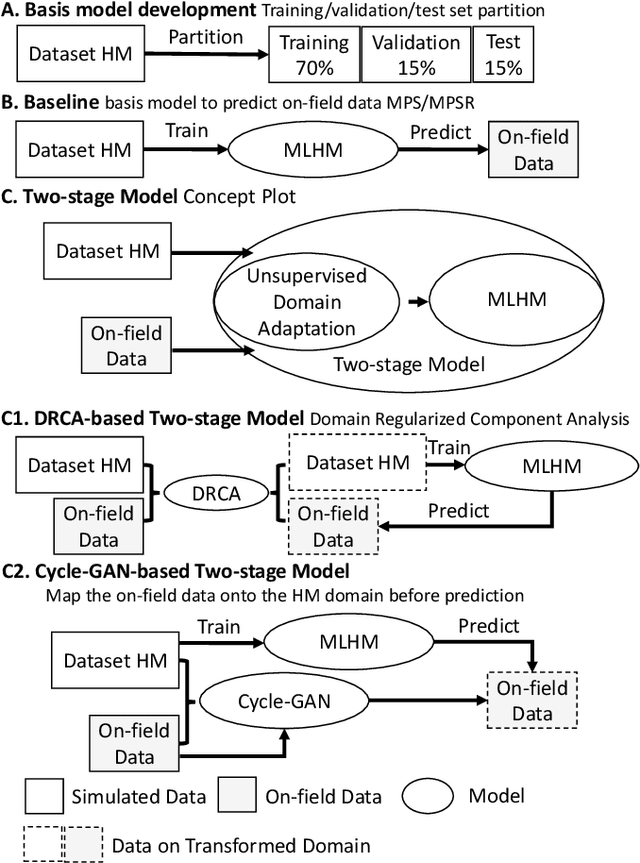
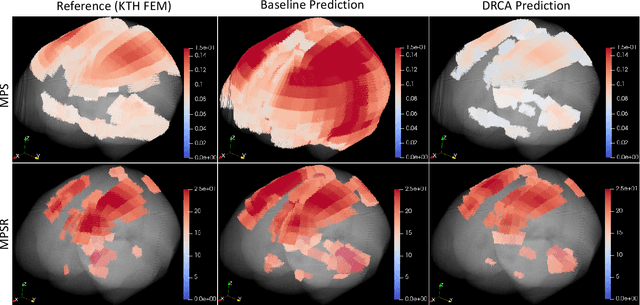
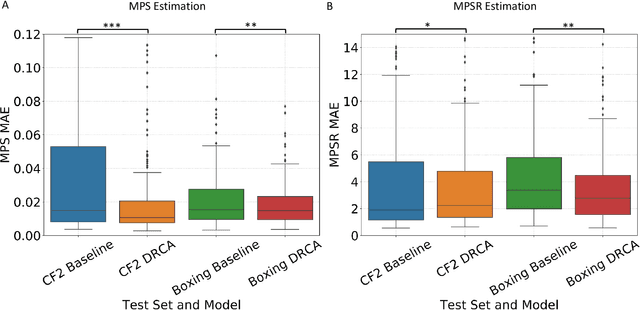
Abstract:Machine learning head models (MLHMs) are developed to estimate brain deformation for early detection of traumatic brain injury (TBI). However, the overfitting to simulated impacts and the lack of generalizability caused by distributional shift of different head impact datasets hinders the broad clinical applications of current MLHMs. We propose brain deformation estimators that integrates unsupervised domain adaptation with a deep neural network to predict whole-brain maximum principal strain (MPS) and MPS rate (MPSR). With 12,780 simulated head impacts, we performed unsupervised domain adaptation on on-field head impacts from 302 college football (CF) impacts and 457 mixed martial arts (MMA) impacts using domain regularized component analysis (DRCA) and cycle-GAN-based methods. The new model improved the MPS/MPSR estimation accuracy, with the DRCA method significantly outperforming other domain adaptation methods in prediction accuracy (p<0.001): MPS RMSE: 0.027 (CF) and 0.037 (MMA); MPSR RMSE: 7.159 (CF) and 13.022 (MMA). On another two hold-out test sets with 195 college football impacts and 260 boxing impacts, the DRCA model significantly outperformed the baseline model without domain adaptation in MPS and MPSR estimation accuracy (p<0.001). The DRCA domain adaptation reduces the MPS/MPSR estimation error to be well below TBI thresholds, enabling accurate brain deformation estimation to detect TBI in future clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge