Thomas Demeester
Rank-Learner: Orthogonal Ranking of Treatment Effects
Feb 03, 2026Abstract:Many decision-making problems require ranking individuals by their treatment effects rather than estimating the exact effect magnitudes. Examples include prioritizing patients for preventive care interventions, or ranking customers by the expected incremental impact of an advertisement. Surprisingly, while causal effect estimation has received substantial attention in the literature, the problem of directly learning rankings of treatment effects has largely remained unexplored. In this paper, we introduce Rank-Learner, a novel two-stage learner that directly learns the ranking of treatment effects from observational data. We first show that naive approaches based on precise treatment effect estimation solve a harder problem than necessary for ranking, while our Rank-Learner optimizes a pairwise learning objective that recovers the true treatment effect ordering, without explicit CATE estimation. We further show that our Rank-Learner is Neyman-orthogonal and thus comes with strong theoretical guarantees, including robustness to estimation errors in the nuisance functions. In addition, our Rank-Learner is model-agnostic, and can be instantiated with arbitrary machine learning models (e.g., neural networks). We demonstrate the effectiveness of our method through extensive experiments where Rank-Learner consistently outperforms standard CATE estimators and non-orthogonal ranking methods. Overall, we provide practitioners with a new, orthogonal two-stage learner for ranking individuals by their treatment effects.
Dynamic Expert-Guided Model Averaging for Causal Discovery
Jan 23, 2026Abstract:Understanding causal relationships is critical for healthcare. Accurate causal models provide a means to enhance the interpretability of predictive models, and furthermore a basis for counterfactual and interventional reasoning and the estimation of treatment effects. However, would-be practitioners of causal discovery face a dizzying array of algorithms without a clear best choice. This abundance of competitive algorithms makes ensembling a natural choice for practical applications. At the same time, real-world use cases frequently face challenges that violate the assumptions of common causal discovery algorithms, forcing heavy reliance on expert knowledge. Inspired by recent work on dynamically requested expert knowledge and LLMs as experts, we present a flexible model averaging method leveraging dynamically requested expert knowledge to ensemble a diverse array of causal discovery algorithms. Experiments demonstrate the efficacy of our method with imperfect experts such as LLMs on both clean and noisy data. We also analyze the impact of different degrees of expert correctness and assess the capabilities of LLMs for clinical causal discovery, providing valuable insights for practitioners.
Hidden State Poisoning Attacks against Mamba-based Language Models
Jan 06, 2026Abstract:State space models (SSMs) like Mamba offer efficient alternatives to Transformer-based language models, with linear time complexity. Yet, their adversarial robustness remains critically unexplored. This paper studies the phenomenon whereby specific short input phrases induce a partial amnesia effect in such models, by irreversibly overwriting information in their hidden states, referred to as a Hidden State Poisoning Attack (HiSPA). Our benchmark RoBench25 allows evaluating a model's information retrieval capabilities when subject to HiSPAs, and confirms the vulnerability of SSMs against such attacks. Even a recent 52B hybrid SSM-Transformer model from the Jamba family collapses on RoBench25 under optimized HiSPA triggers, whereas pure Transformers do not. We also observe that HiSPA triggers significantly weaken the Jamba model on the popular Open-Prompt-Injections benchmark, unlike pure Transformers. Finally, our interpretability study reveals patterns in Mamba's hidden layers during HiSPAs that could be used to build a HiSPA mitigation system. The full code and data to reproduce the experiments can be found at https://anonymous.4open.science/r/hispa_anonymous-5DB0.
Modeling Clinical Uncertainty in Radiology Reports: from Explicit Uncertainty Markers to Implicit Reasoning Pathways
Nov 06, 2025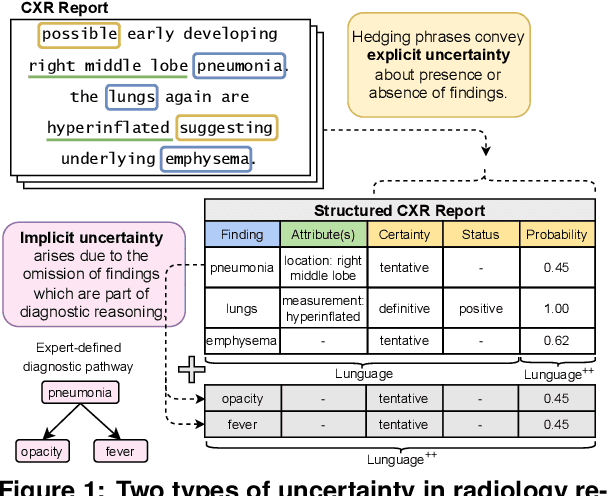
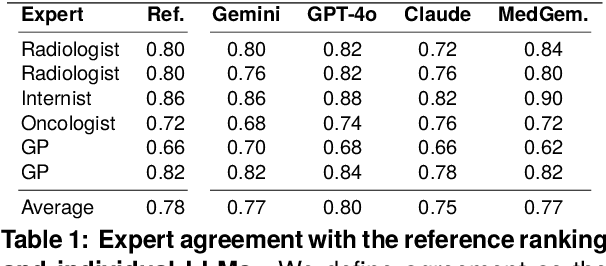
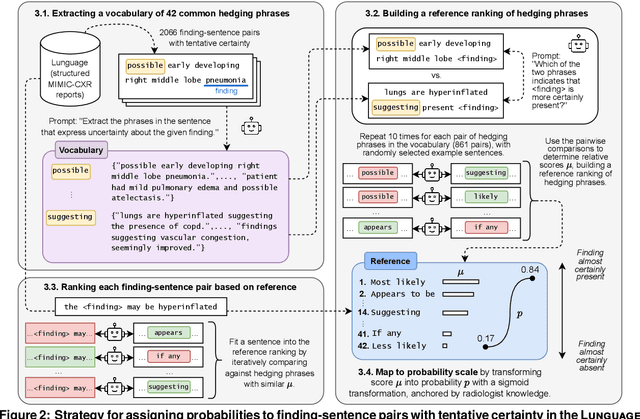
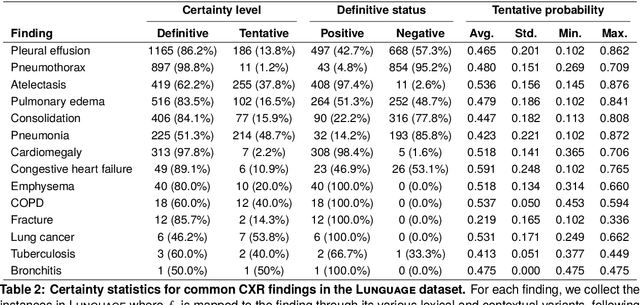
Abstract:Radiology reports are invaluable for clinical decision-making and hold great potential for automated analysis when structured into machine-readable formats. These reports often contain uncertainty, which we categorize into two distinct types: (i) Explicit uncertainty reflects doubt about the presence or absence of findings, conveyed through hedging phrases. These vary in meaning depending on the context, making rule-based systems insufficient to quantify the level of uncertainty for specific findings; (ii) Implicit uncertainty arises when radiologists omit parts of their reasoning, recording only key findings or diagnoses. Here, it is often unclear whether omitted findings are truly absent or simply unmentioned for brevity. We address these challenges with a two-part framework. We quantify explicit uncertainty by creating an expert-validated, LLM-based reference ranking of common hedging phrases, and mapping each finding to a probability value based on this reference. In addition, we model implicit uncertainty through an expansion framework that systematically adds characteristic sub-findings derived from expert-defined diagnostic pathways for 14 common diagnoses. Using these methods, we release Lunguage++, an expanded, uncertainty-aware version of the Lunguage benchmark of fine-grained structured radiology reports. This enriched resource enables uncertainty-aware image classification, faithful diagnostic reasoning, and new investigations into the clinical impact of diagnostic uncertainty.
Personalized Treatment Effect Estimation from Unstructured Data
Jul 28, 2025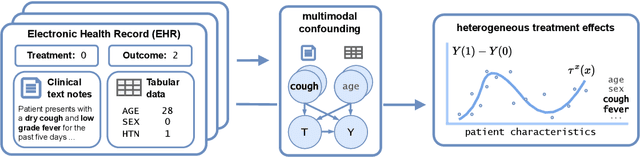
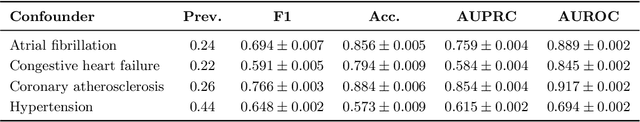
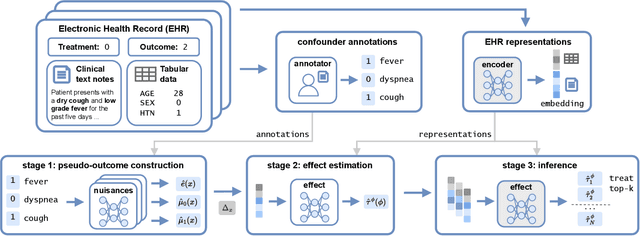
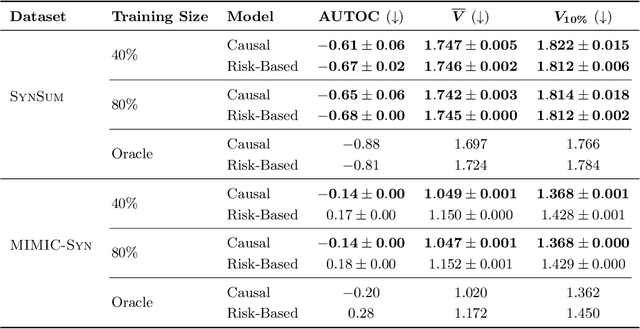
Abstract:Existing methods for estimating personalized treatment effects typically rely on structured covariates, limiting their applicability to unstructured data. Yet, leveraging unstructured data for causal inference has considerable application potential, for instance in healthcare, where clinical notes or medical images are abundant. To this end, we first introduce an approximate 'plug-in' method trained directly on the neural representations of unstructured data. However, when these fail to capture all confounding information, the method may be subject to confounding bias. We therefore introduce two theoretically grounded estimators that leverage structured measurements of the confounders during training, but allow estimating personalized treatment effects purely from unstructured inputs, while avoiding confounding bias. When these structured measurements are only available for a non-representative subset of the data, these estimators may suffer from sampling bias. To address this, we further introduce a regression-based correction that accounts for the non-uniform sampling, assuming the sampling mechanism is known or can be well-estimated. Our experiments on two benchmark datasets show that the plug-in method, directly trainable on large unstructured datasets, achieves strong empirical performance across all settings, despite its simplicity.
Improved Allergy Wheal Detection for the Skin Prick Automated Test Device
Jun 06, 2025Abstract:Background: The skin prick test (SPT) is the gold standard for diagnosing sensitization to inhalant allergies. The Skin Prick Automated Test (SPAT) device was designed for increased consistency in test results, and captures 32 images to be jointly used for allergy wheal detection and delineation, which leads to a diagnosis. Materials and Methods: Using SPAT data from $868$ patients with suspected inhalant allergies, we designed an automated method to detect and delineate wheals on these images. To this end, $10,416$ wheals were manually annotated by drawing detailed polygons along the edges. The unique data-modality of the SPAT device, with $32$ images taken under distinct lighting conditions, requires a custom-made approach. Our proposed method consists of two parts: a neural network component that segments the wheals on the pixel level, followed by an algorithmic and interpretable approach for detecting and delineating the wheals. Results: We evaluate the performance of our method on a hold-out validation set of $217$ patients. As a baseline we use a single conventionally lighted image per SPT as input to our method. Conclusion: Using the $32$ SPAT images under various lighting conditions offers a considerably higher accuracy than a single image in conventional, uniform light.
Efficient Text Encoders for Labor Market Analysis
May 30, 2025Abstract:Labor market analysis relies on extracting insights from job advertisements, which provide valuable yet unstructured information on job titles and corresponding skill requirements. While state-of-the-art methods for skill extraction achieve strong performance, they depend on large language models (LLMs), which are computationally expensive and slow. In this paper, we propose \textbf{ConTeXT-match}, a novel contrastive learning approach with token-level attention that is well-suited for the extreme multi-label classification task of skill classification. \textbf{ConTeXT-match} significantly improves skill extraction efficiency and performance, achieving state-of-the-art results with a lightweight bi-encoder model. To support robust evaluation, we introduce \textbf{Skill-XL}, a new benchmark with exhaustive, sentence-level skill annotations that explicitly address the redundancy in the large label space. Finally, we present \textbf{JobBERT V2}, an improved job title normalization model that leverages extracted skills to produce high-quality job title representations. Experiments demonstrate that our models are efficient, accurate, and scalable, making them ideal for large-scale, real-time labor market analysis.
Error Optimization: Overcoming Exponential Signal Decay in Deep Predictive Coding Networks
May 26, 2025Abstract:Predictive Coding (PC) offers a biologically plausible alternative to backpropagation for neural network training, yet struggles with deeper architectures. This paper identifies the root cause: an inherent signal decay problem where gradients attenuate exponentially with depth, becoming computationally negligible due to numerical precision constraints. To address this fundamental limitation, we introduce Error Optimization (EO), a novel reparameterization that preserves PC's theoretical properties while eliminating signal decay. By optimizing over prediction errors rather than states, EO enables signals to reach all layers simultaneously and without attenuation, converging orders of magnitude faster than standard PC. Experiments across multiple architectures and datasets demonstrate that EO matches backpropagation's performance even for deeper models where conventional PC struggles. Besides practical improvements, our work provides theoretical insight into PC dynamics and establishes a foundation for scaling biologically-inspired learning to deeper architectures on digital hardware and beyond.
Single- vs. Dual-Prompt Dialogue Generation with LLMs for Job Interviews in Human Resources
Feb 25, 2025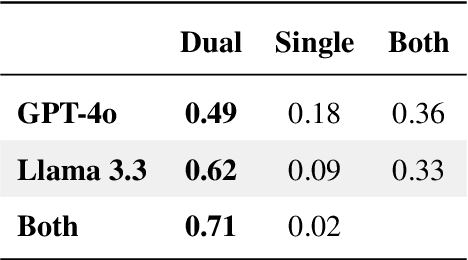
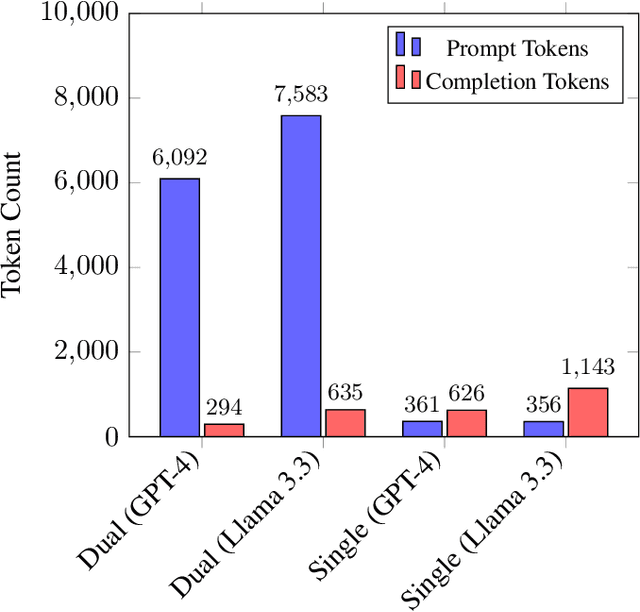
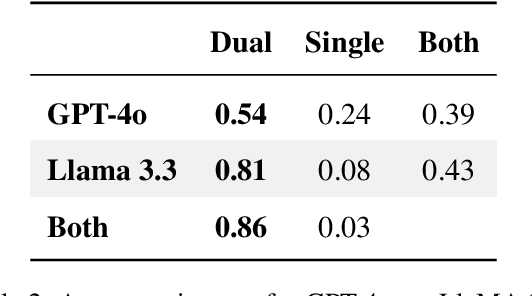
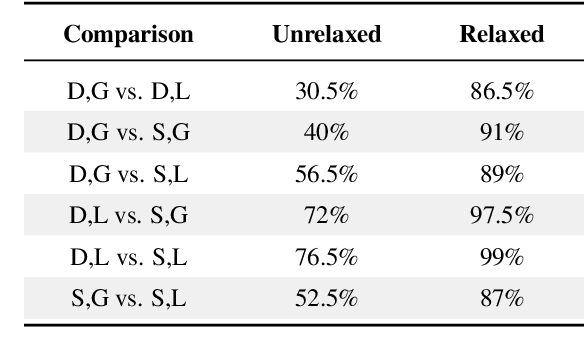
Abstract:Optimizing language models for use in conversational agents requires large quantities of example dialogues. Increasingly, these dialogues are synthetically generated by using powerful large language models (LLMs), especially in domains with challenges to obtain authentic human data. One such domain is human resources (HR). In this context, we compare two LLM-based dialogue generation methods for the use case of generating HR job interviews, and assess whether one method generates higher-quality dialogues that are more challenging to distinguish from genuine human discourse. The first method uses a single prompt to generate the complete interview dialog. The second method uses two agents that converse with each other. To evaluate dialogue quality under each method, we ask a judge LLM to determine whether AI was used for interview generation, using pairwise interview comparisons. We demonstrate that despite a sixfold increase in token cost, interviews generated with the dual-prompt method achieve a win rate up to ten times higher than those generated with the single-prompt method. This difference remains consistent regardless of whether GPT-4o or Llama 3.3 70B is used for either interview generation or judging quality.
Prior Knowledge Injection into Deep Learning Models Predicting Gene Expression from Whole Slide Images
Jan 23, 2025Abstract:Cancer diagnosis and prognosis primarily depend on clinical parameters such as age and tumor grade, and are increasingly complemented by molecular data, such as gene expression, from tumor sequencing. However, sequencing is costly and delays oncology workflows. Recent advances in Deep Learning allow to predict molecular information from morphological features within Whole Slide Images (WSIs), offering a cost-effective proxy of the molecular markers. While promising, current methods lack the robustness to fully replace direct sequencing. Here we aim to improve existing methods by introducing a model-agnostic framework that allows to inject prior knowledge on gene-gene interactions into Deep Learning architectures, thereby increasing accuracy and robustness. We design the framework to be generic and flexibly adaptable to a wide range of architectures. In a case study on breast cancer, our strategy leads to an average increase of 983 significant genes (out of 25,761) across all 18 experiments, with 14 generalizing to an increase on an independent dataset. Our findings reveal a high potential for injection of prior knowledge to increase gene expression prediction performance from WSIs across a wide range of architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge