Paloma Rabaey
Modeling Clinical Uncertainty in Radiology Reports: from Explicit Uncertainty Markers to Implicit Reasoning Pathways
Nov 06, 2025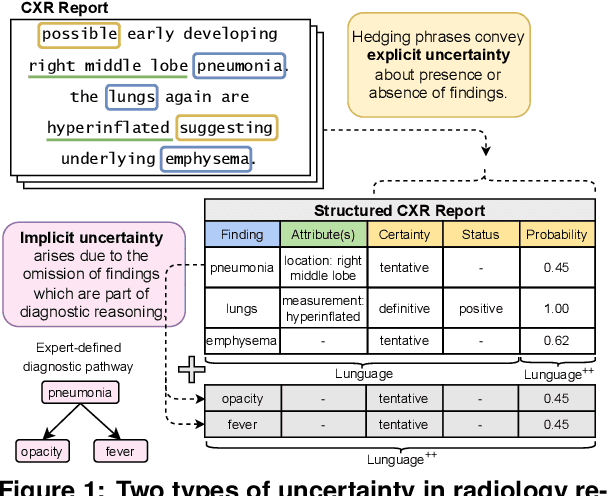
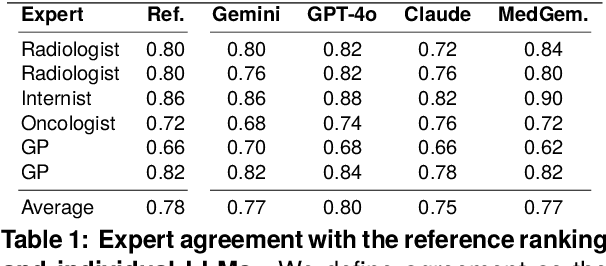
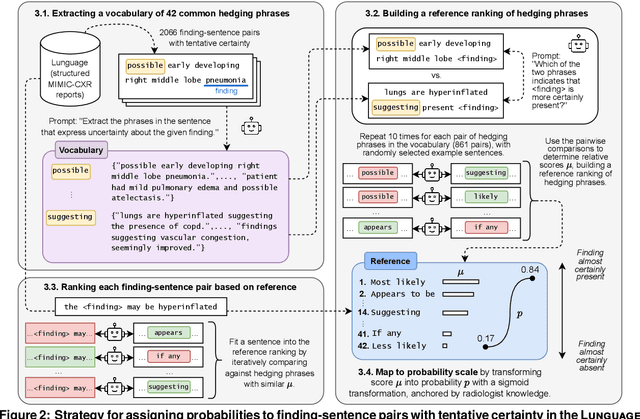
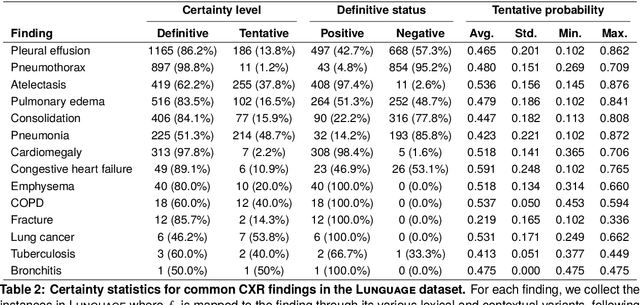
Abstract:Radiology reports are invaluable for clinical decision-making and hold great potential for automated analysis when structured into machine-readable formats. These reports often contain uncertainty, which we categorize into two distinct types: (i) Explicit uncertainty reflects doubt about the presence or absence of findings, conveyed through hedging phrases. These vary in meaning depending on the context, making rule-based systems insufficient to quantify the level of uncertainty for specific findings; (ii) Implicit uncertainty arises when radiologists omit parts of their reasoning, recording only key findings or diagnoses. Here, it is often unclear whether omitted findings are truly absent or simply unmentioned for brevity. We address these challenges with a two-part framework. We quantify explicit uncertainty by creating an expert-validated, LLM-based reference ranking of common hedging phrases, and mapping each finding to a probability value based on this reference. In addition, we model implicit uncertainty through an expansion framework that systematically adds characteristic sub-findings derived from expert-defined diagnostic pathways for 14 common diagnoses. Using these methods, we release Lunguage++, an expanded, uncertainty-aware version of the Lunguage benchmark of fine-grained structured radiology reports. This enriched resource enables uncertainty-aware image classification, faithful diagnostic reasoning, and new investigations into the clinical impact of diagnostic uncertainty.
Lunguage: A Benchmark for Structured and Sequential Chest X-ray Interpretation
May 27, 2025Abstract:Radiology reports convey detailed clinical observations and capture diagnostic reasoning that evolves over time. However, existing evaluation methods are limited to single-report settings and rely on coarse metrics that fail to capture fine-grained clinical semantics and temporal dependencies. We introduce LUNGUAGE,a benchmark dataset for structured radiology report generation that supports both single-report evaluation and longitudinal patient-level assessment across multiple studies. It contains 1,473 annotated chest X-ray reports, each reviewed by experts, and 80 of them contain longitudinal annotations to capture disease progression and inter-study intervals, also reviewed by experts. Using this benchmark, we develop a two-stage framework that transforms generated reports into fine-grained, schema-aligned structured representations, enabling longitudinal interpretation. We also propose LUNGUAGESCORE, an interpretable metric that compares structured outputs at the entity, relation, and attribute level while modeling temporal consistency across patient timelines. These contributions establish the first benchmark dataset, structuring framework, and evaluation metric for sequential radiology reporting, with empirical results demonstrating that LUNGUAGESCORE effectively supports structured report evaluation. The code is available at: https://github.com/SuperSupermoon/Lunguage
Prior Knowledge Injection into Deep Learning Models Predicting Gene Expression from Whole Slide Images
Jan 23, 2025Abstract:Cancer diagnosis and prognosis primarily depend on clinical parameters such as age and tumor grade, and are increasingly complemented by molecular data, such as gene expression, from tumor sequencing. However, sequencing is costly and delays oncology workflows. Recent advances in Deep Learning allow to predict molecular information from morphological features within Whole Slide Images (WSIs), offering a cost-effective proxy of the molecular markers. While promising, current methods lack the robustness to fully replace direct sequencing. Here we aim to improve existing methods by introducing a model-agnostic framework that allows to inject prior knowledge on gene-gene interactions into Deep Learning architectures, thereby increasing accuracy and robustness. We design the framework to be generic and flexibly adaptable to a wide range of architectures. In a case study on breast cancer, our strategy leads to an average increase of 983 significant genes (out of 25,761) across all 18 experiments, with 14 generalizing to an increase on an independent dataset. Our findings reveal a high potential for injection of prior knowledge to increase gene expression prediction performance from WSIs across a wide range of architectures.
Debiasing Synthetic Data Generated by Deep Generative Models
Nov 06, 2024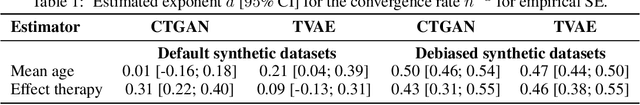
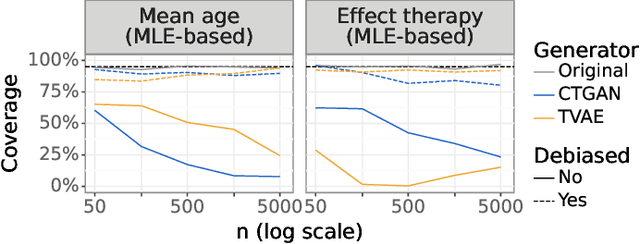
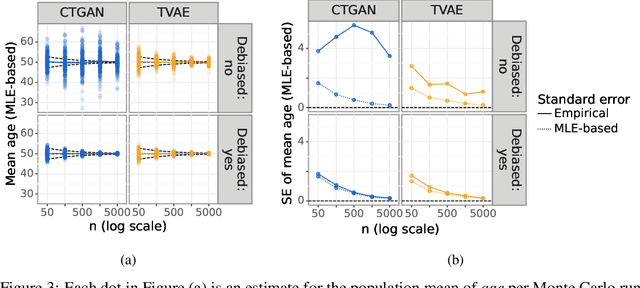
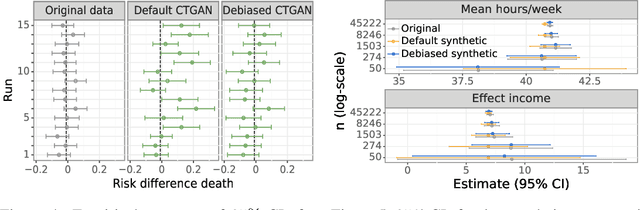
Abstract:While synthetic data hold great promise for privacy protection, their statistical analysis poses significant challenges that necessitate innovative solutions. The use of deep generative models (DGMs) for synthetic data generation is known to induce considerable bias and imprecision into synthetic data analyses, compromising their inferential utility as opposed to original data analyses. This bias and uncertainty can be substantial enough to impede statistical convergence rates, even in seemingly straightforward analyses like mean calculation. The standard errors of such estimators then exhibit slower shrinkage with sample size than the typical 1 over root-$n$ rate. This complicates fundamental calculations like p-values and confidence intervals, with no straightforward remedy currently available. In response to these challenges, we propose a new strategy that targets synthetic data created by DGMs for specific data analyses. Drawing insights from debiased and targeted machine learning, our approach accounts for biases, enhances convergence rates, and facilitates the calculation of estimators with easily approximated large sample variances. We exemplify our proposal through a simulation study on toy data and two case studies on real-world data, highlighting the importance of tailoring DGMs for targeted data analysis. This debiasing strategy contributes to advancing the reliability and applicability of synthetic data in statistical inference.
SynSUM -- Synthetic Benchmark with Structured and Unstructured Medical Records
Sep 13, 2024Abstract:We present the SynSUM benchmark, a synthetic dataset linking unstructured clinical notes to structured background variables. The dataset consists of 10,000 artificial patient records containing tabular variables (like symptoms, diagnoses and underlying conditions) and related notes describing the fictional patient encounter in the domain of respiratory diseases. The tabular portion of the data is generated through a Bayesian network, where both the causal structure between the variables and the conditional probabilities are proposed by an expert based on domain knowledge. We then prompt a large language model (GPT-4o) to generate a clinical note related to this patient encounter, describing the patient symptoms and additional context. The SynSUM dataset is primarily designed to facilitate research on clinical information extraction in the presence of tabular background variables, which can be linked through domain knowledge to concepts of interest to be extracted from the text - the symptoms, in the case of SynSUM. Secondary uses include research on the automation of clinical reasoning over both tabular data and text, causal effect estimation in the presence of tabular and/or textual confounders, and multi-modal synthetic data generation. The dataset can be downloaded from https://github.com/prabaey/SynSUM.
Clinical Reasoning over Tabular Data and Text with Bayesian Networks
Mar 19, 2024Abstract:Bayesian networks are well-suited for clinical reasoning on tabular data, but are less compatible with natural language data, for which neural networks provide a successful framework. This paper compares and discusses strategies to augment Bayesian networks with neural text representations, both in a generative and discriminative manner. This is illustrated with simulation results for a primary care use case (diagnosis of pneumonia) and discussed in a broader clinical context.
Synthetic Data: Can We Trust Statistical Estimators?
Dec 13, 2023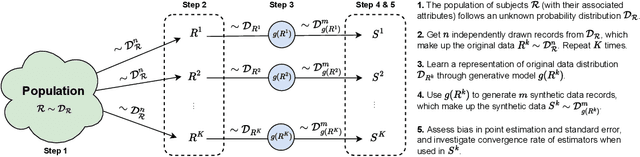
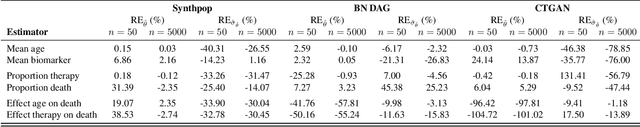
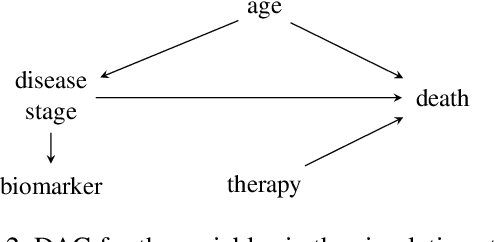
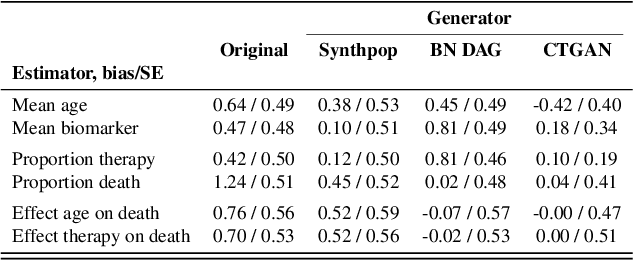
Abstract:The increasing interest in data sharing makes synthetic data appealing. However, the analysis of synthetic data raises a unique set of methodological challenges. In this work, we highlight the importance of inferential utility and provide empirical evidence against naive inference from synthetic data (that handles these as if they were really observed). We argue that the rate of false-positive findings (type 1 error) will be unacceptably high, even when the estimates are unbiased. One of the reasons is the underestimation of the true standard error, which may even progressively increase with larger sample sizes due to slower convergence. This is especially problematic for deep generative models. Before publishing synthetic data, it is essential to develop statistical inference tools for such data.
Neural Bayesian Network Understudy
Nov 15, 2022Abstract:Bayesian Networks may be appealing for clinical decision-making due to their inclusion of causal knowledge, but their practical adoption remains limited as a result of their inability to deal with unstructured data. While neural networks do not have this limitation, they are not interpretable and are inherently unable to deal with causal structure in the input space. Our goal is to build neural networks that combine the advantages of both approaches. Motivated by the perspective to inject causal knowledge while training such neural networks, this work presents initial steps in that direction. We demonstrate how a neural network can be trained to output conditional probabilities, providing approximately the same functionality as a Bayesian Network. Additionally, we propose two training strategies that allow encoding the independence relations inferred from a given causal structure into the neural network. We present initial results in a proof-of-concept setting, showing that the neural model acts as an understudy to its Bayesian Network counterpart, approximating its probabilistic and causal properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge