François Remy
ChocoLlama: Lessons Learned From Teaching Llamas Dutch
Dec 10, 2024Abstract:While Large Language Models (LLMs) have shown remarkable capabilities in natural language understanding and generation, their performance often lags in lower-resource, non-English languages due to biases in the training data. In this work, we explore strategies for adapting the primarily English LLMs (Llama-2 and Llama-3) to Dutch, a language spoken by 30 million people worldwide yet often underrepresented in LLM development. We collect 104GB of Dutch text ($32$B tokens) from various sources to first apply continued pretraining using low-rank adaptation (LoRA), complemented with Dutch posttraining strategies provided by prior work. For Llama-2, we consider using (i) the tokenizer of the original model, and (ii) training a new, Dutch-specific tokenizer combined with embedding reinitialization. We evaluate our adapted models, ChocoLlama-2, both on standard benchmarks and a novel Dutch benchmark, ChocoLlama-Bench. Our results demonstrate that LoRA can effectively scale for language adaptation, and that tokenizer modification with careful weight reinitialization can improve performance. Notably, Llama-3 was released during the course of this project and, upon evaluation, demonstrated superior Dutch capabilities compared to our Dutch-adapted versions of Llama-2. We hence apply the same adaptation technique to Llama-3, using its original tokenizer. While our adaptation methods enhanced Llama-2's Dutch capabilities, we found limited gains when applying the same techniques to Llama-3. This suggests that for ever improving, multilingual foundation models, language adaptation techniques may benefit more from focusing on language-specific posttraining rather than on continued pretraining. We hope this work contributes to the broader understanding of adapting LLMs to lower-resource languages, and to the development of Dutch LLMs in particular.
Trans-Tokenization and Cross-lingual Vocabulary Transfers: Language Adaptation of LLMs for Low-Resource NLP
Aug 08, 2024Abstract:The development of monolingual language models for low and mid-resource languages continues to be hindered by the difficulty in sourcing high-quality training data. In this study, we present a novel cross-lingual vocabulary transfer strategy, trans-tokenization, designed to tackle this challenge and enable more efficient language adaptation. Our approach focuses on adapting a high-resource monolingual LLM to an unseen target language by initializing the token embeddings of the target language using a weighted average of semantically similar token embeddings from the source language. For this, we leverage a translation resource covering both the source and target languages. We validate our method with the Tweeties, a series of trans-tokenized LLMs, and demonstrate their competitive performance on various downstream tasks across a small but diverse set of languages. Additionally, we introduce Hydra LLMs, models with multiple swappable language modeling heads and embedding tables, which further extend the capabilities of our trans-tokenization strategy. By designing a Hydra LLM based on the multilingual model TowerInstruct, we developed a state-of-the-art machine translation model for Tatar, in a zero-shot manner, completely bypassing the need for high-quality parallel data. This breakthrough is particularly significant for low-resource languages like Tatar, where high-quality parallel data is hard to come by. By lowering the data and time requirements for training high-quality models, our trans-tokenization strategy allows for the development of LLMs for a wider range of languages, especially those with limited resources. We hope that our work will inspire further research and collaboration in the field of cross-lingual vocabulary transfer and contribute to the empowerment of languages on a global scale.
In-Context Learning for Extreme Multi-Label Classification
Jan 22, 2024Abstract:Multi-label classification problems with thousands of classes are hard to solve with in-context learning alone, as language models (LMs) might lack prior knowledge about the precise classes or how to assign them, and it is generally infeasible to demonstrate every class in a prompt. We propose a general program, $\texttt{Infer--Retrieve--Rank}$, that defines multi-step interactions between LMs and retrievers to efficiently tackle such problems. We implement this program using the $\texttt{DSPy}$ programming model, which specifies in-context systems in a declarative manner, and use $\texttt{DSPy}$ optimizers to tune it towards specific datasets by bootstrapping only tens of few-shot examples. Our primary extreme classification program, optimized separately for each task, attains state-of-the-art results across three benchmarks (HOUSE, TECH, TECHWOLF). We apply the same program to a benchmark with vastly different characteristics and attain competitive performance as well (BioDEX). Unlike prior work, our proposed solution requires no finetuning, is easily applicable to new tasks, alleviates prompt engineering, and requires only tens of labeled examples. Our code is public at https://github.com/KarelDO/xmc.dspy.
BioLORD-2023: Semantic Textual Representations Fusing LLM and Clinical Knowledge Graph Insights
Nov 27, 2023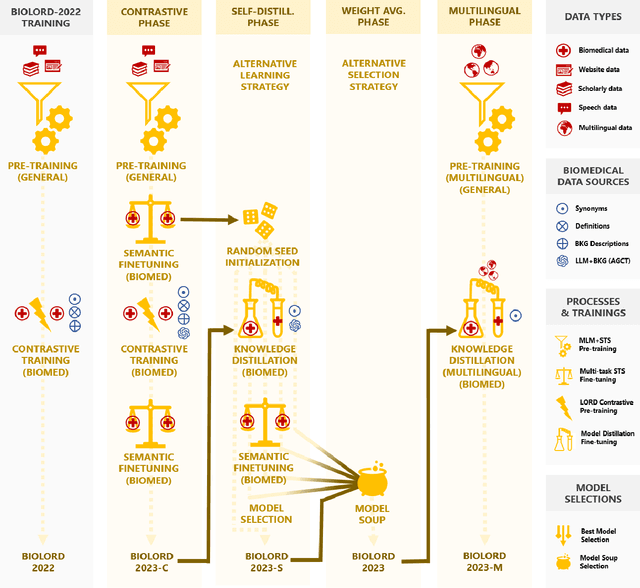
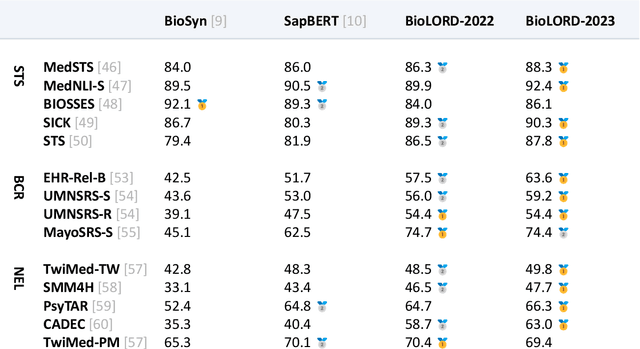
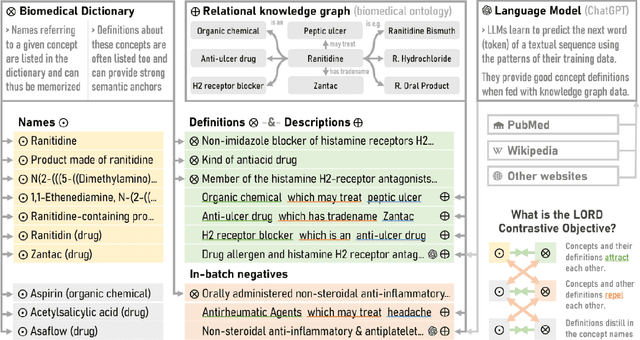
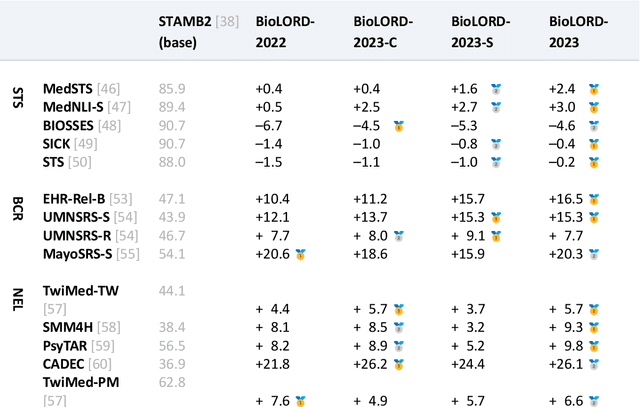
Abstract:In this study, we investigate the potential of Large Language Models to complement biomedical knowledge graphs in the training of semantic models for the biomedical and clinical domains. Drawing on the wealth of the UMLS knowledge graph and harnessing cutting-edge Large Language Models, we propose a new state-of-the-art approach for obtaining high-fidelity representations of biomedical concepts and sentences, consisting of three steps: an improved contrastive learning phase, a novel self-distillation phase, and a weight averaging phase. Through rigorous evaluations via the extensive BioLORD testing suite and diverse downstream tasks, we demonstrate consistent and substantial performance improvements over the previous state of the art (e.g. +2pts on MedSTS, +2.5pts on MedNLI-S, +6.1pts on EHR-Rel-B). Besides our new state-of-the-art biomedical model for English, we also distill and release a multilingual model compatible with 50+ languages and finetuned on 7 European languages. Many clinical pipelines can benefit from our latest models. Our new multilingual model enables a range of languages to benefit from our advancements in biomedical semantic representation learning, opening a new avenue for bioinformatics researchers around the world. As a result, we hope to see BioLORD-2023 becoming a precious tool for future biomedical applications.
Tik-to-Tok: Translating Language Models One Token at a Time: An Embedding Initialization Strategy for Efficient Language Adaptation
Oct 05, 2023



Abstract:Training monolingual language models for low and mid-resource languages is made challenging by limited and often inadequate pretraining data. In this study, we propose a novel model conversion strategy to address this issue, adapting high-resources monolingual language models to a new target language. By generalizing over a word translation dictionary encompassing both the source and target languages, we map tokens from the target tokenizer to semantically similar tokens from the source language tokenizer. This one-to-many token mapping improves tremendously the initialization of the embedding table for the target language. We conduct experiments to convert high-resource models to mid- and low-resource languages, namely Dutch and Frisian. These converted models achieve a new state-of-the-art performance on these languages across all sorts of downstream tasks. By reducing significantly the amount of data and time required for training state-of-the-art models, our novel model conversion strategy has the potential to benefit many languages worldwide.
Boosting Adverse Drug Event Normalization on Social Media: General-Purpose Model Initialization and Biomedical Semantic Text Similarity Benefit Zero-Shot Linking in Informal Contexts
Jul 31, 2023


Abstract:Biomedical entity linking, also known as biomedical concept normalization, has recently witnessed the rise to prominence of zero-shot contrastive models. However, the pre-training material used for these models has, until now, largely consisted of specialist biomedical content such as MIMIC-III clinical notes (Johnson et al., 2016) and PubMed papers (Sayers et al., 2021; Gao et al., 2020). While the resulting in-domain models have shown promising results for many biomedical tasks, adverse drug event normalization on social media texts has so far remained challenging for them (Portelli et al., 2022). In this paper, we propose a new approach for adverse drug event normalization on social media relying on general-purpose model initialization via BioLORD (Remy et al., 2022) and a semantic-text-similarity fine-tuning named STS. Our experimental results on several social media datasets demonstrate the effectiveness of our proposed approach, by achieving state-of-the-art performance. Based on its strong performance across all the tested datasets, we believe this work could emerge as a turning point for the task of adverse drug event normalization on social media and has the potential to serve as a benchmark for future research in the field.
Automatic Glossary of Clinical Terminology: a Large-Scale Dictionary of Biomedical Definitions Generated from Ontological Knowledge
Jun 01, 2023Abstract:Background: More than 400,000 biomedical concepts and some of their relationships are contained in SnomedCT, a comprehensive biomedical ontology. However, their concept names are not always readily interpretable by non-experts, or patients looking at their own electronic health records (EHR). Clear definitions or descriptions in understandable language are often not available. Therefore, generating human-readable definitions for biomedical concepts might help make the information they encode more accessible and understandable to a wider public. Objective: In this article, we introduce the Automatic Glossary of Clinical Terminology (AGCT), a large-scale biomedical dictionary of clinical concepts generated using high-quality information extracted from the biomedical knowledge contained in SnomedCT. Methods: We generate a novel definition for every SnomedCT concept, after prompting the OpenAI Turbo model, a variant of GPT 3.5, using a high-quality verbalization of the SnomedCT relationships of the to-be-defined concept. A significant subset of the generated definitions was subsequently judged by NLP researchers with biomedical expertise on 5-point scales along the following three axes: factuality, insight, and fluency. Results: AGCT contains 422,070 computer-generated definitions for SnomedCT concepts, covering various domains such as diseases, procedures, drugs, and anatomy. The average length of the definitions is 49 words. The definitions were assigned average scores of over 4.5 out of 5 on all three axes, indicating a majority of factual, insightful, and fluent definitions. Conclusion: AGCT is a novel and valuable resource for biomedical tasks that require human-readable definitions for SnomedCT concepts. It can also serve as a base for developing robust biomedical retrieval models or other applications that leverage natural language understanding of biomedical knowledge.
BioDEX: Large-Scale Biomedical Adverse Drug Event Extraction for Real-World Pharmacovigilance
May 22, 2023



Abstract:Timely and accurate extraction of Adverse Drug Events (ADE) from biomedical literature is paramount for public safety, but involves slow and costly manual labor. We set out to improve drug safety monitoring (pharmacovigilance, PV) through the use of Natural Language Processing (NLP). We introduce BioDEX, a large-scale resource for Biomedical adverse Drug Event Extraction, rooted in the historical output of drug safety reporting in the U.S. BioDEX consists of 65k abstracts and 19k full-text biomedical papers with 256k associated document-level safety reports created by medical experts. The core features of these reports include the reported weight, age, and biological sex of a patient, a set of drugs taken by the patient, the drug dosages, the reactions experienced, and whether the reaction was life threatening. In this work, we consider the task of predicting the core information of the report given its originating paper. We estimate human performance to be 72.0% F1, whereas our best model achieves 62.3% F1, indicating significant headroom on this task. We also begin to explore ways in which these models could help professional PV reviewers. Our code and data are available: https://github.com/KarelDO/BioDEX.
Detecting Idiomatic Multiword Expressions in Clinical Terminology using Definition-Based Representation Learning
May 11, 2023Abstract:This paper shines a light on the potential of definition-based semantic models for detecting idiomatic and semi-idiomatic multiword expressions (MWEs) in clinical terminology. Our study focuses on biomedical entities defined in the UMLS ontology and aims to help prioritize the translation efforts of these entities. In particular, we develop an effective tool for scoring the idiomaticity of biomedical MWEs based on the degree of similarity between the semantic representations of those MWEs and a weighted average of the representation of their constituents. We achieve this using a biomedical language model trained to produce similar representations for entity names and their definitions, called BioLORD. The importance of this definition-based approach is highlighted by comparing the BioLORD model to two other state-of-the-art biomedical language models based on Transformer: SapBERT and CODER. Our results show that the BioLORD model has a strong ability to identify idiomatic MWEs, not replicated in other models. Our corpus-free idiomaticity estimation helps ontology translators to focus on more challenging MWEs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge