Simone Scaboro
Boosting Adverse Drug Event Normalization on Social Media: General-Purpose Model Initialization and Biomedical Semantic Text Similarity Benefit Zero-Shot Linking in Informal Contexts
Jul 31, 2023


Abstract:Biomedical entity linking, also known as biomedical concept normalization, has recently witnessed the rise to prominence of zero-shot contrastive models. However, the pre-training material used for these models has, until now, largely consisted of specialist biomedical content such as MIMIC-III clinical notes (Johnson et al., 2016) and PubMed papers (Sayers et al., 2021; Gao et al., 2020). While the resulting in-domain models have shown promising results for many biomedical tasks, adverse drug event normalization on social media texts has so far remained challenging for them (Portelli et al., 2022). In this paper, we propose a new approach for adverse drug event normalization on social media relying on general-purpose model initialization via BioLORD (Remy et al., 2022) and a semantic-text-similarity fine-tuning named STS. Our experimental results on several social media datasets demonstrate the effectiveness of our proposed approach, by achieving state-of-the-art performance. Based on its strong performance across all the tested datasets, we believe this work could emerge as a turning point for the task of adverse drug event normalization on social media and has the potential to serve as a benchmark for future research in the field.
Extensive Evaluation of Transformer-based Architectures for Adverse Drug Events Extraction
Jun 08, 2023



Abstract:Adverse Event (ADE) extraction is one of the core tasks in digital pharmacovigilance, especially when applied to informal texts. This task has been addressed by the Natural Language Processing community using large pre-trained language models, such as BERT. Despite the great number of Transformer-based architectures used in the literature, it is unclear which of them has better performances and why. Therefore, in this paper we perform an extensive evaluation and analysis of 19 Transformer-based models for ADE extraction on informal texts. We compare the performance of all the considered models on two datasets with increasing levels of informality (forums posts and tweets). We also combine the purely Transformer-based models with two commonly-used additional processing layers (CRF and LSTM), and analyze their effect on the models performance. Furthermore, we use a well-established feature importance technique (SHAP) to correlate the performance of the models with a set of features that describe them: model category (AutoEncoding, AutoRegressive, Text-to-Text), pretraining domain, training from scratch, and model size in number of parameters. At the end of our analyses, we identify a list of take-home messages that can be derived from the experimental data.
AILAB-Udine@SMM4H 22: Limits of Transformers and BERT Ensembles
Sep 07, 2022
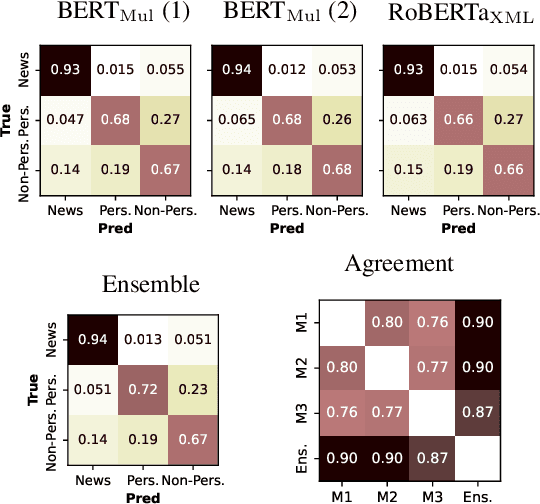

Abstract:This paper describes the models developed by the AILAB-Udine team for the SMM4H 22 Shared Task. We explored the limits of Transformer based models on text classification, entity extraction and entity normalization, tackling Tasks 1, 2, 5, 6 and 10. The main take-aways we got from participating in different tasks are: the overwhelming positive effects of combining different architectures when using ensemble learning, and the great potential of generative models for term normalization.
Increasing Adverse Drug Events extraction robustness on social media: case study on negation and speculation
Sep 06, 2022



Abstract:In the last decade, an increasing number of users have started reporting Adverse Drug Events (ADE) on social media platforms, blogs, and health forums. Given the large volume of reports, pharmacovigilance has focused on ways to use Natural Language Processing (NLP) techniques to rapidly examine these large collections of text, detecting mentions of drug-related adverse reactions to trigger medical investigations. However, despite the growing interest in the task and the advances in NLP, the robustness of these models in face of linguistic phenomena such as negations and speculations is an open research question. Negations and speculations are pervasive phenomena in natural language, and can severely hamper the ability of an automated system to discriminate between factual and nonfactual statements in text. In this paper we take into consideration four state-of-the-art systems for ADE detection on social media texts. We introduce SNAX, a benchmark to test their performance against samples containing negated and speculated ADEs, showing their fragility against these phenomena. We then introduce two possible strategies to increase the robustness of these models, showing that both of them bring significant increases in performance, lowering the number of spurious entities predicted by the models by 60% for negation and 80% for speculations.
NADE: A Benchmark for Robust Adverse Drug Events Extraction in Face of Negations
Sep 24, 2021
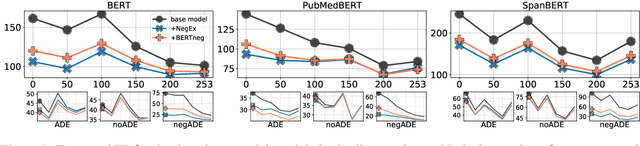


Abstract:Adverse Drug Event (ADE) extraction models can rapidly examine large collections of social media texts, detecting mentions of drug-related adverse reactions and trigger medical investigations. However, despite the recent advances in NLP, it is currently unknown if such models are robust in face of negation, which is pervasive across language varieties. In this paper we evaluate three state-of-the-art systems, showing their fragility against negation, and then we introduce two possible strategies to increase the robustness of these models: a pipeline approach, relying on a specific component for negation detection; an augmentation of an ADE extraction dataset to artificially create negated samples and further train the models. We show that both strategies bring significant increases in performance, lowering the number of spurious entities predicted by the models. Our dataset and code will be publicly released to encourage research on the topic.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge