Simon Ellershaw
BioDEX: Large-Scale Biomedical Adverse Drug Event Extraction for Real-World Pharmacovigilance
May 22, 2023



Abstract:Timely and accurate extraction of Adverse Drug Events (ADE) from biomedical literature is paramount for public safety, but involves slow and costly manual labor. We set out to improve drug safety monitoring (pharmacovigilance, PV) through the use of Natural Language Processing (NLP). We introduce BioDEX, a large-scale resource for Biomedical adverse Drug Event Extraction, rooted in the historical output of drug safety reporting in the U.S. BioDEX consists of 65k abstracts and 19k full-text biomedical papers with 256k associated document-level safety reports created by medical experts. The core features of these reports include the reported weight, age, and biological sex of a patient, a set of drugs taken by the patient, the drug dosages, the reactions experienced, and whether the reaction was life threatening. In this work, we consider the task of predicting the core information of the report given its originating paper. We estimate human performance to be 72.0% F1, whereas our best model achieves 62.3% F1, indicating significant headroom on this task. We also begin to explore ways in which these models could help professional PV reviewers. Our code and data are available: https://github.com/KarelDO/BioDEX.
Unsupervised Human Pose Estimation through Transforming Shape Templates
May 10, 2021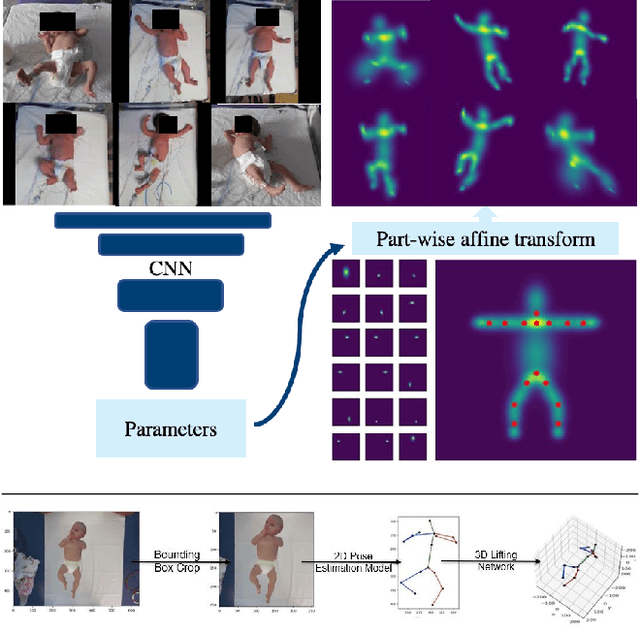
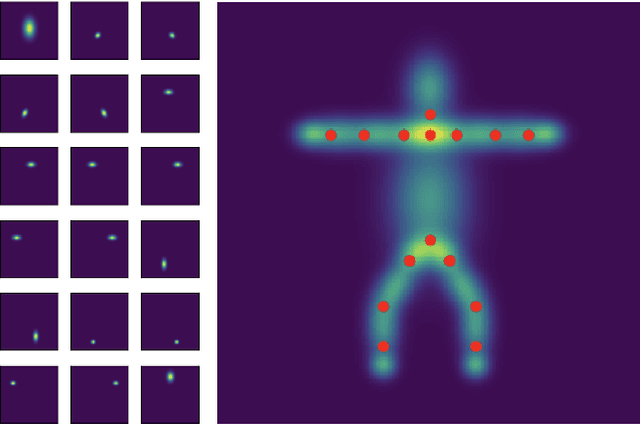
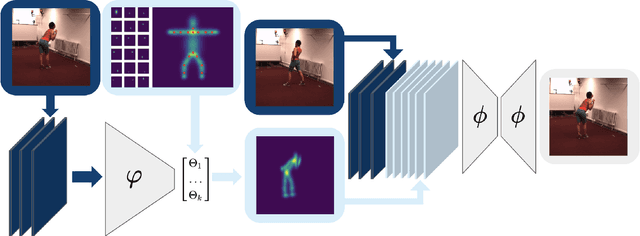
Abstract:Human pose estimation is a major computer vision problem with applications ranging from augmented reality and video capture to surveillance and movement tracking. In the medical context, the latter may be an important biomarker for neurological impairments in infants. Whilst many methods exist, their application has been limited by the need for well annotated large datasets and the inability to generalize to humans of different shapes and body compositions, e.g. children and infants. In this paper we present a novel method for learning pose estimators for human adults and infants in an unsupervised fashion. We approach this as a learnable template matching problem facilitated by deep feature extractors. Human-interpretable landmarks are estimated by transforming a template consisting of predefined body parts that are characterized by 2D Gaussian distributions. Enforcing a connectivity prior guides our model to meaningful human shape representations. We demonstrate the effectiveness of our approach on two different datasets including adults and infants.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge