Idoia Ochoa
Interpretable Causal Representation Learning for Biological Data in the Pathway Space
Jun 14, 2025Abstract:Predicting the impact of genomic and drug perturbations in cellular function is crucial for understanding gene functions and drug effects, ultimately leading to improved therapies. To this end, Causal Representation Learning (CRL) constitutes one of the most promising approaches, as it aims to identify the latent factors that causally govern biological systems, thus facilitating the prediction of the effect of unseen perturbations. Yet, current CRL methods fail in reconciling their principled latent representations with known biological processes, leading to models that are not interpretable. To address this major issue, we present SENA-discrepancy-VAE, a model based on the recently proposed CRL method discrepancy-VAE, that produces representations where each latent factor can be interpreted as the (linear) combination of the activity of a (learned) set of biological processes. To this extent, we present an encoder, SENA-{\delta}, that efficiently compute and map biological processes' activity levels to the latent causal factors. We show that SENA-discrepancy-VAE achieves predictive performances on unseen combinations of interventions that are comparable with its original, non-interpretable counterpart, while inferring causal latent factors that are biologically meaningful.
Towards a more inductive world for drug repurposing approaches
Nov 24, 2023Abstract:Drug-target interaction (DTI) prediction is a challenging, albeit essential task in drug repurposing. Learning on graph models have drawn special attention as they can significantly reduce drug repurposing costs and time commitment. However, many current approaches require high-demanding additional information besides DTIs that complicates their evaluation process and usability. Additionally, structural differences in the learning architecture of current models hinder their fair benchmarking. In this work, we first perform an in-depth evaluation of current DTI datasets and prediction models through a robust benchmarking process, and show that DTI prediction methods based on transductive models lack generalization and lead to inflated performance when evaluated as previously done in the literature, hence not being suited for drug repurposing approaches. We then propose a novel biologically-driven strategy for negative edge subsampling and show through in vitro validation that newly discovered interactions are indeed true. We envision this work as the underpinning for future fair benchmarking and robust model design. All generated resources and tools are publicly available as a python package.
DZip: improved general-purpose lossless compression based on novel neural network modeling
Nov 08, 2019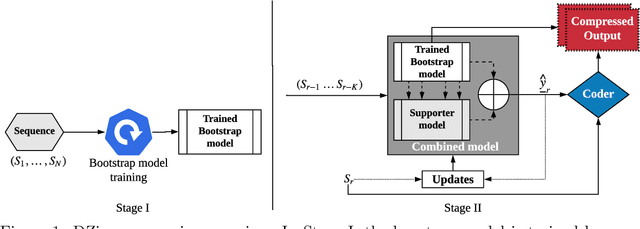

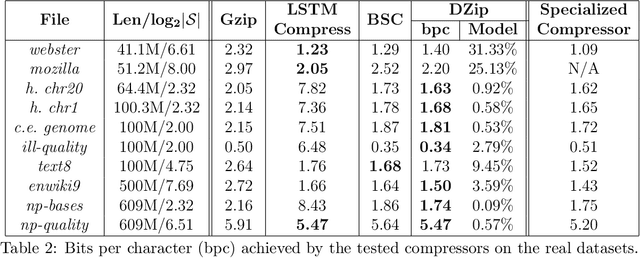
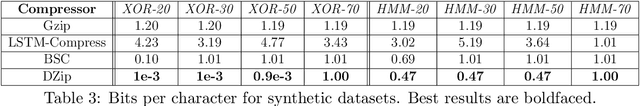
Abstract:We consider lossless compression based on statistical data modeling followed by prediction-based encoding, where an accurate statistical model for the input data leads to substantial improvements in compression. We propose DZip, a general-purpose compressor for sequential data that exploits the well-known modeling capabilities of neural networks (NNs) for prediction, followed by arithmetic coding. Dzip uses a novel hybrid architecture based on adaptive and semi-adaptive training. Unlike most NN based compressors, DZip does not require additional training data and is not restricted to specific data types, only needing the alphabet size of the input data. The proposed compressor outperforms general-purpose compressors such as Gzip (on average 26% reduction) on a variety of real datasets, achieves near-optimal compression on synthetic datasets, and performs close to specialized compressors for large sequence lengths, without any human input. The main limitation of DZip in its current implementation is the encoding/decoding time, which limits its practicality. Nevertheless, the results showcase the potential of developing improved general-purpose compressors based on neural networks and hybrid modeling.
DeepZip: Lossless Data Compression using Recurrent Neural Networks
Nov 20, 2018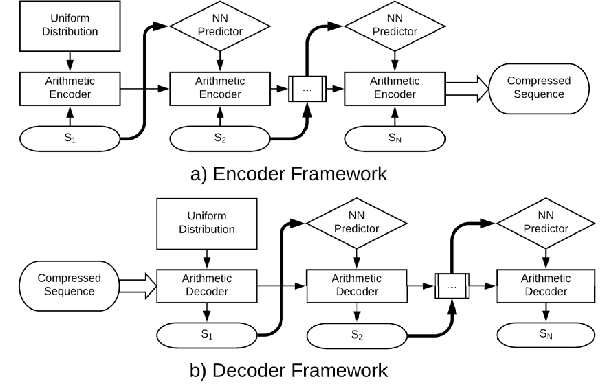
Abstract:Sequential data is being generated at an unprecedented pace in various forms, including text and genomic data. This creates the need for efficient compression mechanisms to enable better storage, transmission and processing of such data. To solve this problem, many of the existing compressors attempt to learn models for the data and perform prediction-based compression. Since neural networks are known as universal function approximators with the capability to learn arbitrarily complex mappings, and in practice show excellent performance in prediction tasks, we explore and devise methods to compress sequential data using neural network predictors. We combine recurrent neural network predictors with an arithmetic coder and losslessly compress a variety of synthetic, text and genomic datasets. The proposed compressor outperforms Gzip on the real datasets and achieves near-optimal compression for the synthetic datasets. The results also help understand why and where neural networks are good alternatives for traditional finite context models
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge