Comparisons of Graph Neural Networks on Cancer Classification Leveraging a Joint of Phenotypic and Genetic Features
Paper and Code
Jan 14, 2021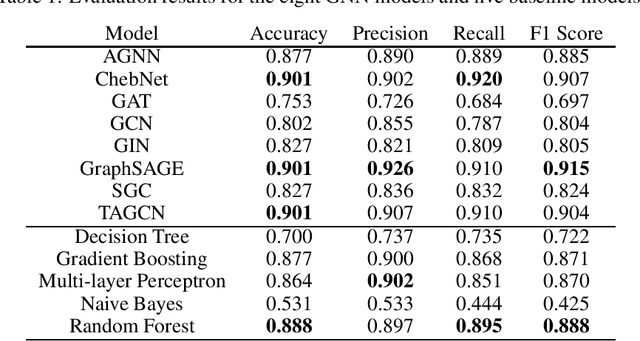
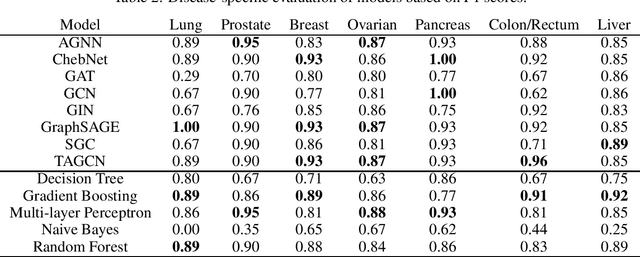
Cancer is responsible for millions of deaths worldwide every year. Although significant progress hasbeen achieved in cancer medicine, many issues remain to be addressed for improving cancer therapy.Appropriate cancer patient stratification is the prerequisite for selecting appropriate treatment plan, ascancer patients are of known heterogeneous genetic make-ups and phenotypic differences. In thisstudy, built upon deep phenotypic characterizations extractable from Mayo Clinic electronic healthrecords (EHRs) and genetic test reports for a collection of cancer patients, we evaluated variousgraph neural networks (GNNs) leveraging a joint of phenotypic and genetic features for cancer typeclassification. Models were applied and fine-tuned on the Mayo Clinic cancer disease dataset. Theassessment was done through the reported accuracy, precision, recall, and F1 values as well as throughF1 scores based on the disease class. Per our evaluation results, GNNs on average outperformed thebaseline models with mean statistics always being higher that those of the baseline models (0.849 vs0.772 for accuracy, 0.858 vs 0.794 for precision, 0.843 vs 0.759 for recall, and 0.843 vs 0.855 for F1score). Among GNNs, ChebNet, GraphSAGE, and TAGCN showed the best performance, while GATshowed the worst. We applied and compared eight GNN models including AGNN, ChebNet, GAT,GCN, GIN, GraphSAGE, SGC, and TAGCN on the Mayo Clinic cancer disease dataset and assessedtheir performance as well as compared them with each other and with more conventional machinelearning models such as decision tree, gradient boosting, multi-layer perceptron, naive bayes, andrandom forest which we used as the baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge