Jinlian Wang
Explainable Diagnosis Prediction through Neuro-Symbolic Integration
Oct 01, 2024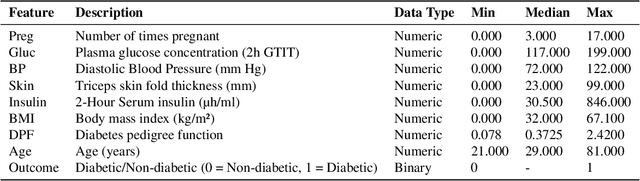

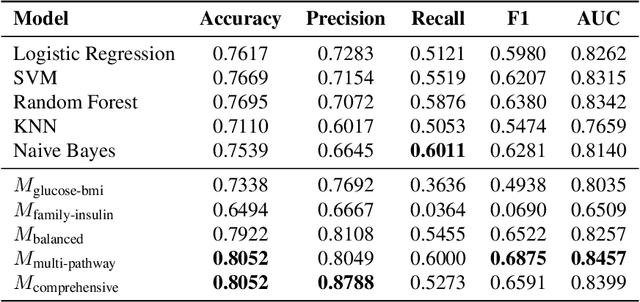
Abstract:Diagnosis prediction is a critical task in healthcare, where timely and accurate identification of medical conditions can significantly impact patient outcomes. Traditional machine learning and deep learning models have achieved notable success in this domain but often lack interpretability which is a crucial requirement in clinical settings. In this study, we explore the use of neuro-symbolic methods, specifically Logical Neural Networks (LNNs), to develop explainable models for diagnosis prediction. Essentially, we design and implement LNN-based models that integrate domain-specific knowledge through logical rules with learnable thresholds. Our models, particularly $M_{\text{multi-pathway}}$ and $M_{\text{comprehensive}}$, demonstrate superior performance over traditional models such as Logistic Regression, SVM, and Random Forest, achieving higher accuracy (up to 80.52\%) and AUROC scores (up to 0.8457) in the case study of diabetes prediction. The learned weights and thresholds within the LNN models provide direct insights into feature contributions, enhancing interpretability without compromising predictive power. These findings highlight the potential of neuro-symbolic approaches in bridging the gap between accuracy and explainability in healthcare AI applications. By offering transparent and adaptable diagnostic models, our work contributes to the advancement of precision medicine and supports the development of equitable healthcare solutions. Future research will focus on extending these methods to larger and more diverse datasets to further validate their applicability across different medical conditions and populations.
Large Language Models Struggle in Token-Level Clinical Named Entity Recognition
Jun 30, 2024Abstract:Large Language Models (LLMs) have revolutionized various sectors, including healthcare where they are employed in diverse applications. Their utility is particularly significant in the context of rare diseases, where data scarcity, complexity, and specificity pose considerable challenges. In the clinical domain, Named Entity Recognition (NER) stands out as an essential task and it plays a crucial role in extracting relevant information from clinical texts. Despite the promise of LLMs, current research mostly concentrates on document-level NER, identifying entities in a more general context across entire documents, without extracting their precise location. Additionally, efforts have been directed towards adapting ChatGPT for token-level NER. However, there is a significant research gap when it comes to employing token-level NER for clinical texts, especially with the use of local open-source LLMs. This study aims to bridge this gap by investigating the effectiveness of both proprietary and local LLMs in token-level clinical NER. Essentially, we delve into the capabilities of these models through a series of experiments involving zero-shot prompting, few-shot prompting, retrieval-augmented generation (RAG), and instruction-fine-tuning. Our exploration reveals the inherent challenges LLMs face in token-level NER, particularly in the context of rare diseases, and suggests possible improvements for their application in healthcare. This research contributes to narrowing a significant gap in healthcare informatics and offers insights that could lead to a more refined application of LLMs in the healthcare sector.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge