Anne Blaes
CancerLLM: A Large Language Model in Cancer Domain
Jun 15, 2024



Abstract:Medical Large Language Models (LLMs) such as ClinicalCamel 70B, Llama3-OpenBioLLM 70B have demonstrated impressive performance on a wide variety of medical NLP task.However, there still lacks a large language model (LLM) specifically designed for cancer domain. Moreover, these LLMs typically have billions of parameters, making them computationally expensive for healthcare systems.Thus, in this study, we propose CancerLLM, a model with 7 billion parameters and a Mistral-style architecture, pre-trained on 2,676,642 clinical notes and 515,524 pathology reports covering 17 cancer types, followed by fine-tuning on three cancer-relevant tasks, including cancer phenotypes extraction, cancer diagnosis generation, and cancer treatment plan generation. Our evaluation demonstrated that CancerLLM achieves state-of-the-art results compared to other existing LLMs, with an average F1 score improvement of 8.1\%. Additionally, CancerLLM outperforms other models on two proposed robustness testbeds. This illustrates that CancerLLM can be effectively applied to clinical AI systems, enhancing clinical research and healthcare delivery in the field of cancer.
A Cross-institutional Evaluation on Breast Cancer Phenotyping NLP Algorithms on Electronic Health Records
Mar 15, 2023Abstract:Objective: The generalizability of clinical large language models is usually ignored during the model development process. This study evaluated the generalizability of BERT-based clinical NLP models across different clinical settings through a breast cancer phenotype extraction task. Materials and Methods: Two clinical corpora of breast cancer patients were collected from the electronic health records from the University of Minnesota and the Mayo Clinic, and annotated following the same guideline. We developed three types of NLP models (i.e., conditional random field, bi-directional long short-term memory and CancerBERT) to extract cancer phenotypes from clinical texts. The models were evaluated for their generalizability on different test sets with different learning strategies (model transfer vs. locally trained). The entity coverage score was assessed with their association with the model performances. Results: We manually annotated 200 and 161 clinical documents at UMN and MC, respectively. The corpora of the two institutes were found to have higher similarity between the target entities than the overall corpora. The CancerBERT models obtained the best performances among the independent test sets from two clinical institutes and the permutation test set. The CancerBERT model developed in one institute and further fine-tuned in another institute achieved reasonable performance compared to the model developed on local data (micro-F1: 0.925 vs 0.932). Conclusions: The results indicate the CancerBERT model has the best learning ability and generalizability among the three types of clinical NLP models. The generalizability of the models was found to be correlated with the similarity of the target entities between the corpora.
Predicting Cancer Treatments Induced Cardiotoxicity of Breast Cancer Patients
Jan 31, 2022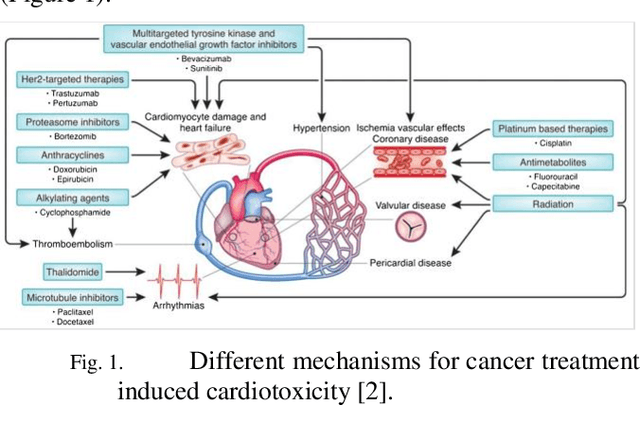
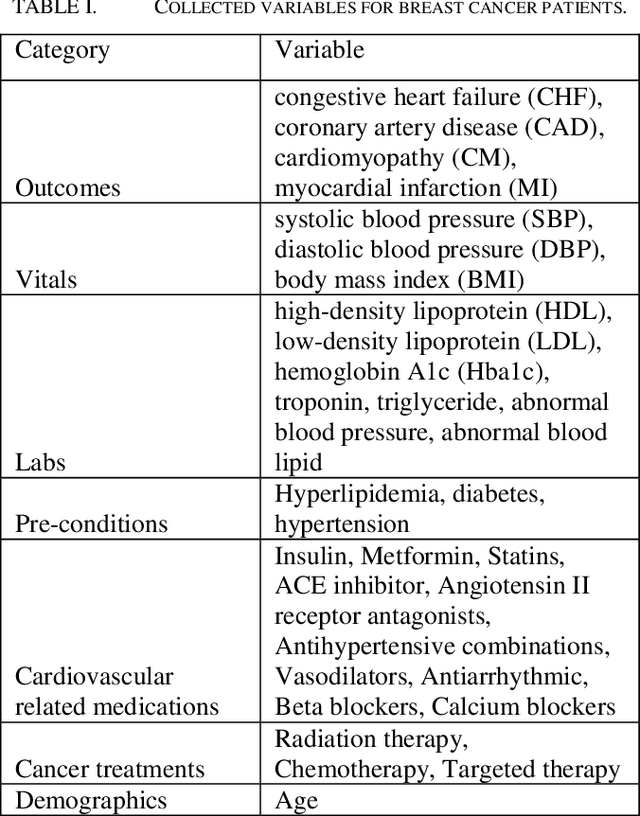
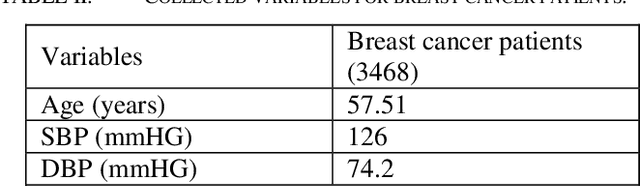
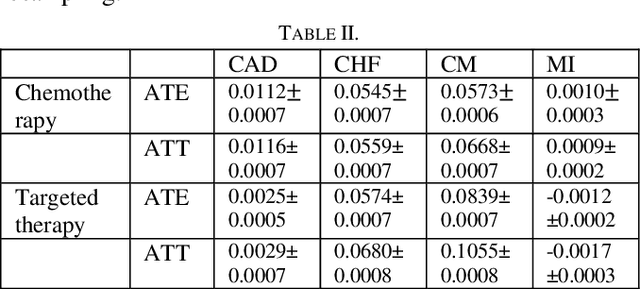
Abstract:Cardiotoxicity induced by the breast cancer treatments (i.e., chemotherapy, targeted therapy and radiation therapy) is a significant problem for breast cancer patients. The cardiotoxicity risk for breast cancer patients receiving different treatments remains unclear. We developed and evaluated risk predictive models for cardiotoxicity in breast cancer patients using EHR data. The AUC scores to predict the CHF, CAD, CM and MI are 0.846, 0.857, 0.858 and 0.804 respectively. After adjusting for baseline differences in cardiovascular health, patients who received chemotherapy or targeted therapy appeared to have higher risk of cardiotoxicity than patients who received radiation therapy. Due to differences in baseline cardiac health across the different breast cancer treatment groups, caution is recommended in interpreting the cardiotoxic effect of these treatments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge