Sicheng Zhou
SimRPD: Optimizing Recruitment Proactive Dialogue Agents through Simulator-Based Data Evaluation and Selection
Jan 08, 2026Abstract:Task-oriented proactive dialogue agents play a pivotal role in recruitment, particularly for steering conversations towards specific business outcomes, such as acquiring social-media contacts for private-channel conversion. Although supervised fine-tuning and reinforcement learning have proven effective for training such agents, their performance is heavily constrained by the scarcity of high-quality, goal-oriented domain-specific training data. To address this challenge, we propose SimRPD, a three-stage framework for training recruitment proactive dialogue agents. First, we develop a high-fidelity user simulator to synthesize large-scale conversational data through multi-turn online dialogue. Then we introduce a multi-dimensional evaluation framework based on Chain-of-Intention (CoI) to comprehensively assess the simulator and effectively select high-quality data, incorporating both global-level and instance-level metrics. Finally, we train the recruitment proactive dialogue agent on the selected dataset. Experiments in a real-world recruitment scenario demonstrate that SimRPD outperforms existing simulator-based data selection strategies, highlighting its practical value for industrial deployment and its potential applicability to other business-oriented dialogue scenarios.
StableToolBench-MirrorAPI: Modeling Tool Environments as Mirrors of 7,000+ Real-World APIs
Mar 26, 2025



Abstract:The rapid advancement of large language models (LLMs) has spurred significant interest in tool learning, where LLMs are augmented with external tools to tackle complex tasks. However, existing tool environments face challenges in balancing stability, scalability, and realness, particularly for benchmarking purposes. To address this problem, we propose MirrorAPI, a novel framework that trains specialized LLMs to accurately simulate real API responses, effectively acting as "mirrors" to tool environments. Using a comprehensive dataset of request-response pairs from 7,000+ APIs, we employ supervised fine-tuning and chain-of-thought reasoning to enhance simulation fidelity. MirrorAPI achieves superior accuracy and stability compared to state-of-the-art methods, as demonstrated by its performance on the newly constructed MirrorAPI-Bench and its integration into StableToolBench.
VidEgoThink: Assessing Egocentric Video Understanding Capabilities for Embodied AI
Oct 15, 2024Abstract:Recent advancements in Multi-modal Large Language Models (MLLMs) have opened new avenues for applications in Embodied AI. Building on previous work, EgoThink, we introduce VidEgoThink, a comprehensive benchmark for evaluating egocentric video understanding capabilities. To bridge the gap between MLLMs and low-level control in Embodied AI, we design four key interrelated tasks: video question-answering, hierarchy planning, visual grounding and reward modeling. To minimize manual annotation costs, we develop an automatic data generation pipeline based on the Ego4D dataset, leveraging the prior knowledge and multimodal capabilities of GPT-4o. Three human annotators then filter the generated data to ensure diversity and quality, resulting in the VidEgoThink benchmark. We conduct extensive experiments with three types of models: API-based MLLMs, open-source image-based MLLMs, and open-source video-based MLLMs. Experimental results indicate that all MLLMs, including GPT-4o, perform poorly across all tasks related to egocentric video understanding. These findings suggest that foundation models still require significant advancements to be effectively applied to first-person scenarios in Embodied AI. In conclusion, VidEgoThink reflects a research trend towards employing MLLMs for egocentric vision, akin to human capabilities, enabling active observation and interaction in the complex real-world environments.
A Review of Reinforcement Learning for Natural Language Processing, and Applications in Healthcare
Oct 23, 2023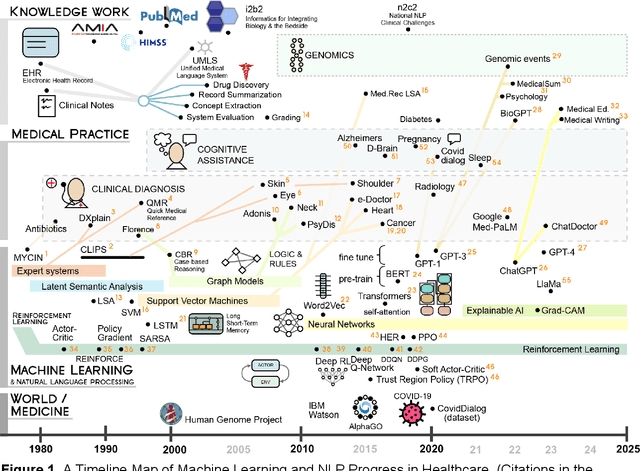

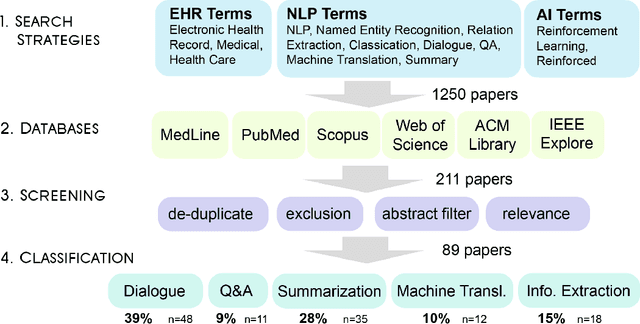
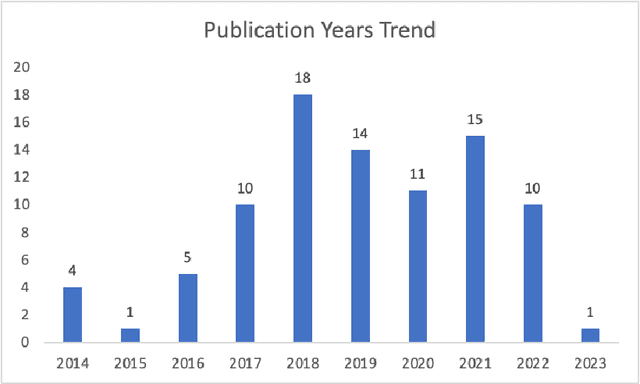
Abstract:Reinforcement learning (RL) has emerged as a powerful approach for tackling complex medical decision-making problems such as treatment planning, personalized medicine, and optimizing the scheduling of surgeries and appointments. It has gained significant attention in the field of Natural Language Processing (NLP) due to its ability to learn optimal strategies for tasks such as dialogue systems, machine translation, and question-answering. This paper presents a review of the RL techniques in NLP, highlighting key advancements, challenges, and applications in healthcare. The review begins by visualizing a roadmap of machine learning and its applications in healthcare. And then it explores the integration of RL with NLP tasks. We examined dialogue systems where RL enables the learning of conversational strategies, RL-based machine translation models, question-answering systems, text summarization, and information extraction. Additionally, ethical considerations and biases in RL-NLP systems are addressed.
A Cross-institutional Evaluation on Breast Cancer Phenotyping NLP Algorithms on Electronic Health Records
Mar 15, 2023Abstract:Objective: The generalizability of clinical large language models is usually ignored during the model development process. This study evaluated the generalizability of BERT-based clinical NLP models across different clinical settings through a breast cancer phenotype extraction task. Materials and Methods: Two clinical corpora of breast cancer patients were collected from the electronic health records from the University of Minnesota and the Mayo Clinic, and annotated following the same guideline. We developed three types of NLP models (i.e., conditional random field, bi-directional long short-term memory and CancerBERT) to extract cancer phenotypes from clinical texts. The models were evaluated for their generalizability on different test sets with different learning strategies (model transfer vs. locally trained). The entity coverage score was assessed with their association with the model performances. Results: We manually annotated 200 and 161 clinical documents at UMN and MC, respectively. The corpora of the two institutes were found to have higher similarity between the target entities than the overall corpora. The CancerBERT models obtained the best performances among the independent test sets from two clinical institutes and the permutation test set. The CancerBERT model developed in one institute and further fine-tuned in another institute achieved reasonable performance compared to the model developed on local data (micro-F1: 0.925 vs 0.932). Conclusions: The results indicate the CancerBERT model has the best learning ability and generalizability among the three types of clinical NLP models. The generalizability of the models was found to be correlated with the similarity of the target entities between the corpora.
Predicting Cancer Treatments Induced Cardiotoxicity of Breast Cancer Patients
Jan 31, 2022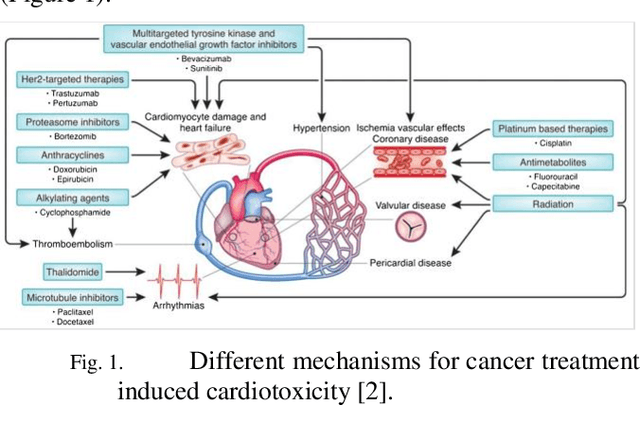
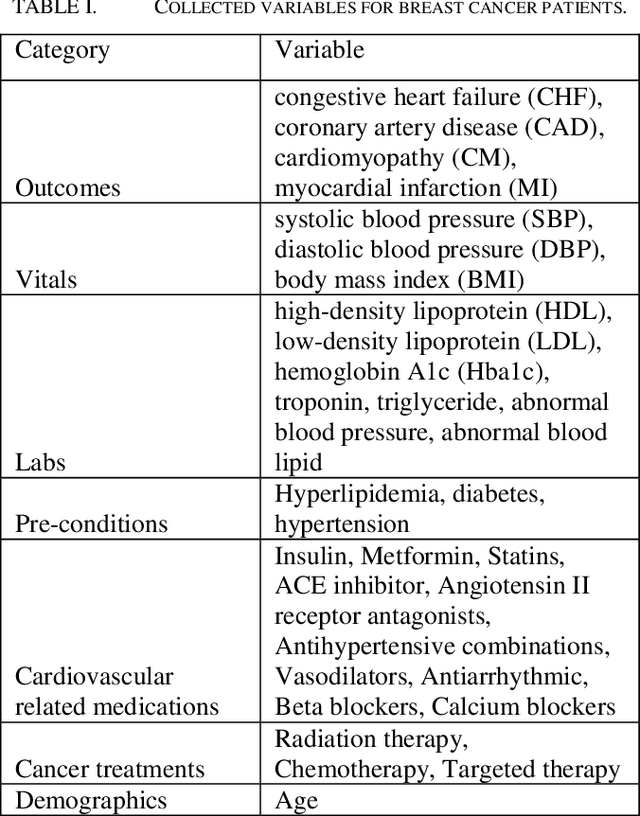
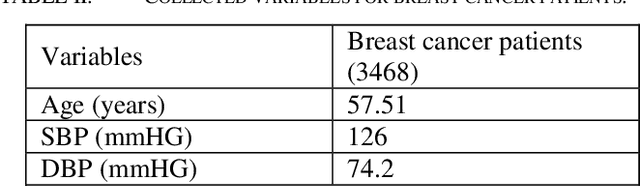
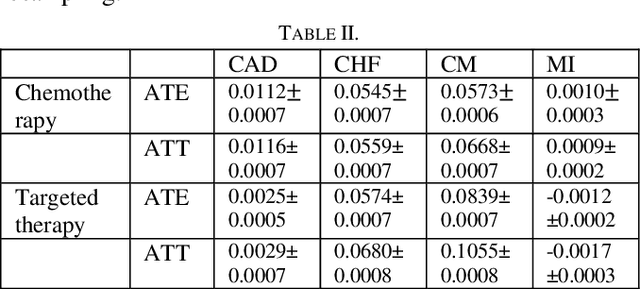
Abstract:Cardiotoxicity induced by the breast cancer treatments (i.e., chemotherapy, targeted therapy and radiation therapy) is a significant problem for breast cancer patients. The cardiotoxicity risk for breast cancer patients receiving different treatments remains unclear. We developed and evaluated risk predictive models for cardiotoxicity in breast cancer patients using EHR data. The AUC scores to predict the CHF, CAD, CM and MI are 0.846, 0.857, 0.858 and 0.804 respectively. After adjusting for baseline differences in cardiovascular health, patients who received chemotherapy or targeted therapy appeared to have higher risk of cardiotoxicity than patients who received radiation therapy. Due to differences in baseline cardiac health across the different breast cancer treatment groups, caution is recommended in interpreting the cardiotoxic effect of these treatments.
CancerBERT: a BERT model for Extracting Breast Cancer Phenotypes from Electronic Health Records
Aug 25, 2021



Abstract:Accurate extraction of breast cancer patients' phenotypes is important for clinical decision support and clinical research. Current models do not take full advantage of cancer domain-specific corpus, whether pre-training Bidirectional Encoder Representations from Transformer model on cancer-specific corpus could improve the performances of extracting breast cancer phenotypes from texts data remains to be explored. The objective of this study is to develop and evaluate the CancerBERT model for extracting breast cancer phenotypes from clinical texts in electronic health records. This data used in the study included 21,291 breast cancer patients diagnosed from 2010 to 2020, patients' clinical notes and pathology reports were collected from the University of Minnesota Clinical Data Repository (UMN). Results: About 3 million clinical notes and pathology reports in electronic health records for 21,291 breast cancer patients were collected to train the CancerBERT model. 200 pathology reports and 50 clinical notes of breast cancer patients that contain 9,685 sentences and 221,356 tokens were manually annotated by two annotators. 20% of the annotated data was used as a test set. Our CancerBERT model achieved the best performance with macro F1 scores equal to 0.876 (95% CI, 0.896-0.902) for exact match and 0.904 (95% CI, 0.896-0.902) for the lenient match. The NER models we developed would facilitate the automated information extraction from clinical texts to further help clinical decision support. Conclusions and Relevance: In this study, we focused on the breast cancer-related concepts extraction from EHR data and obtained a comprehensive annotated dataset that contains 7 types of breast cancer-related concepts. The CancerBERT model with customized vocabulary could significantly improve the performance for extracting breast cancer phenotypes from clinical texts.
Deep Learning Approaches for Extracting Adverse Events and Indications of Dietary Supplements from Clinical Text
Sep 16, 2020


Abstract:The objective of our work is to demonstrate the feasibility of utilizing deep learning models to extract safety signals related to the use of dietary supplements (DS) in clinical text. Two tasks were performed in this study. For the named entity recognition (NER) task, Bi-LSTM-CRF (Bidirectional Long-Short-Term-Memory Conditional Random Fields) and BERT (Bidirectional Encoder Representations from Transformers) models were trained and compared with CRF model as a baseline to recognize the named entities of DS and Events from clinical notes. In the relation extraction (RE) task, two deep learning models, including attention-based Bi-LSTM and CNN (Convolutional Neural Network), and a random forest model were trained to extract the relations between DS and Events, which were categorized into three classes: positive (i.e., indication), negative (i.e., adverse events), and not related. The best performed NER and RE models were further applied on clinical notes mentioning 88 DS for discovering DS adverse events and indications, which were compared with a DS knowledge base. For the NER task, deep learning models achieved a better performance than CRF, with F1 scores above 0.860. The attention-based Bi-LSTM model performed the best in the relation extraction task, with the F1 score of 0.893. When comparing DS event pairs generated by the deep learning models with the knowledge base for DS and Event, we found both known and unknown pairs. Deep learning models can detect adverse events and indication of DS in clinical notes, which hold great potential for monitoring the safety of DS use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge