Bhavani Singh Agnikula Kshatriya
Joy
Neural Language Models with Distant Supervision to Identify Major Depressive Disorder from Clinical Notes
Apr 19, 2021
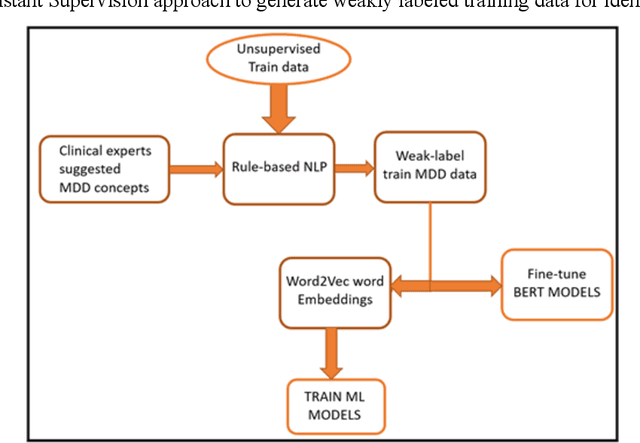
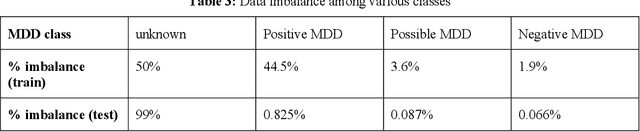
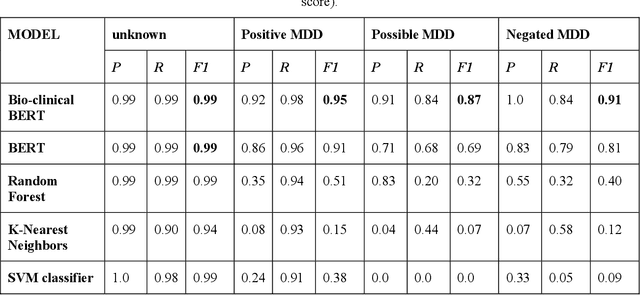
Abstract:Major depressive disorder (MDD) is a prevalent psychiatric disorder that is associated with significant healthcare burden worldwide. Phenotyping of MDD can help early diagnosis and consequently may have significant advantages in patient management. In prior research MDD phenotypes have been extracted from structured Electronic Health Records (EHR) or using Electroencephalographic (EEG) data with traditional machine learning models to predict MDD phenotypes. However, MDD phenotypic information is also documented in free-text EHR data, such as clinical notes. While clinical notes may provide more accurate phenotyping information, natural language processing (NLP) algorithms must be developed to abstract such information. Recent advancements in NLP resulted in state-of-the-art neural language models, such as Bidirectional Encoder Representations for Transformers (BERT) model, which is a transformer-based model that can be pre-trained from a corpus of unsupervised text data and then fine-tuned on specific tasks. However, such neural language models have been underutilized in clinical NLP tasks due to the lack of large training datasets. In the literature, researchers have utilized the distant supervision paradigm to train machine learning models on clinical text classification tasks to mitigate the issue of lacking annotated training data. It is still unknown whether the paradigm is effective for neural language models. In this paper, we propose to leverage the neural language models in a distant supervision paradigm to identify MDD phenotypes from clinical notes. The experimental results indicate that our proposed approach is effective in identifying MDD phenotypes and that the Bio- Clinical BERT, a specific BERT model for clinical data, achieved the best performance in comparison with conventional machine learning models.
Social determinants of health in the era of artificial intelligence with electronic health records: A systematic review
Jan 22, 2021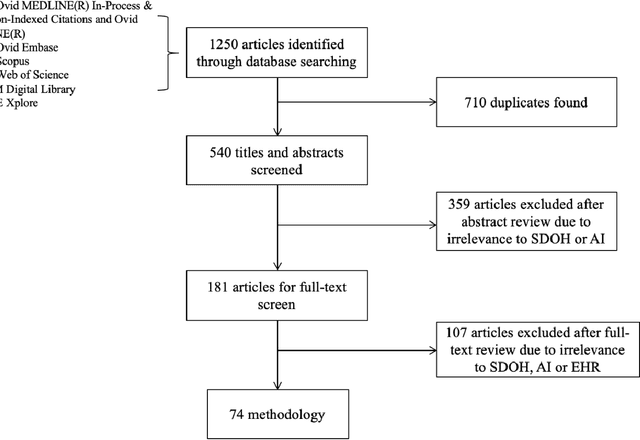
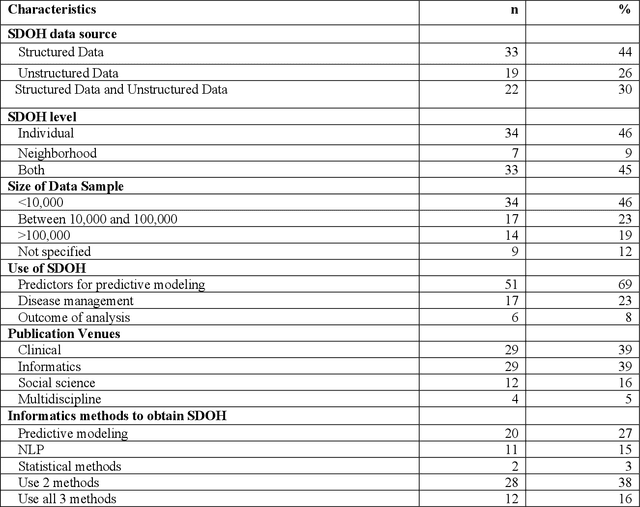
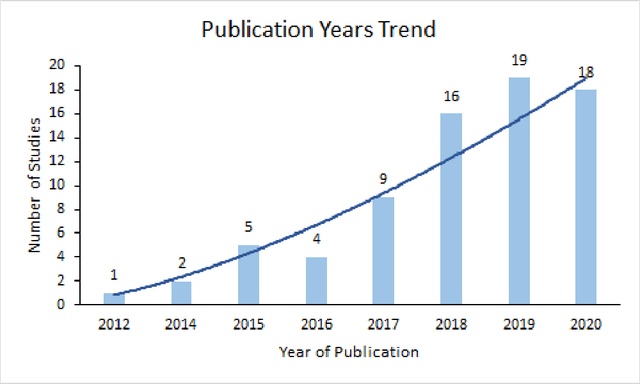
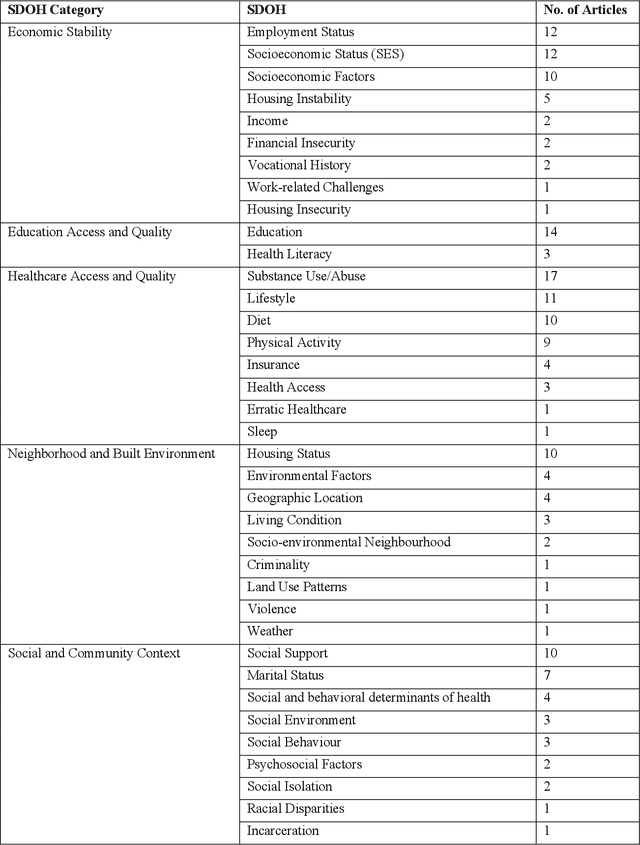
Abstract:There is growing evidence showing the significant role of social determinant of health (SDOH) on a wide variety of health outcomes. In the era of artificial intelligence (AI), electronic health records (EHRs) have been widely used to conduct observational studies. However, how to make the best of SDOH information from EHRs is yet to be studied. In this paper, we systematically reviewed recently published papers and provided a methodology review of AI methods using the SDOH information in EHR data. A total of 1250 articles were retrieved from the literature between 2010 and 2020, and 74 papers were included in this review after abstract and full-text screening. We summarized these papers in terms of general characteristics (including publication years, venues, countries etc.), SDOH types, disease areas, study outcomes, AI methods to extract SDOH from EHRs and AI methods using SDOH for healthcare outcomes. Finally, we conclude this paper with discussion on the current trends, challenges, and future directions on using SDOH from EHRs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge