Anusha Bompelli
Joy
Extracting Lifestyle Factors for Alzheimer's Disease from Clinical Notes Using Deep Learning with Weak Supervision
Jan 25, 2021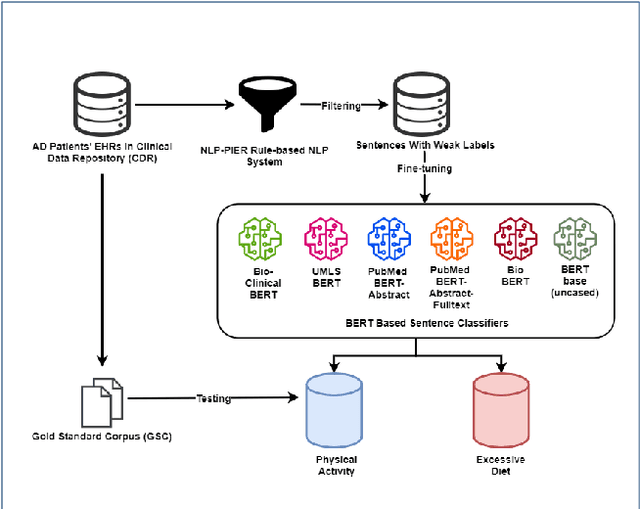
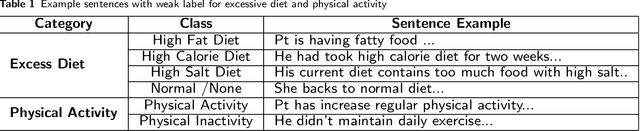
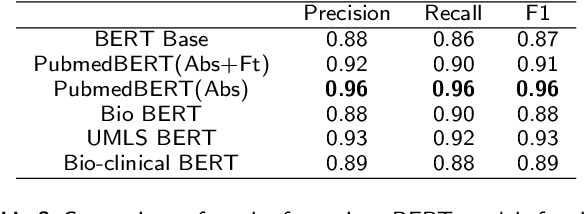
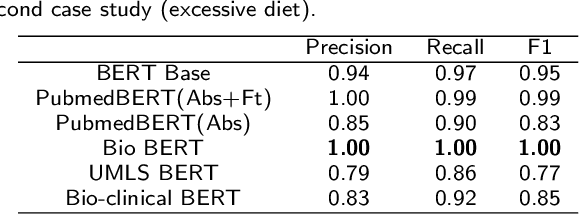
Abstract:Since no effective therapies exist for Alzheimer's disease (AD), prevention has become more critical through lifestyle factor changes and interventions. Analyzing electronic health records (EHR) of patients with AD can help us better understand lifestyle's effect on AD. However, lifestyle information is typically stored in clinical narratives. Thus, the objective of the study was to demonstrate the feasibility of natural language processing (NLP) models to classify lifestyle factors (e.g., physical activity and excessive diet) from clinical texts. We automatically generated labels for the training data by using a rule-based NLP algorithm. We conducted weak supervision for pre-trained Bidirectional Encoder Representations from Transformers (BERT) models on the weakly labeled training corpus. These models include the BERT base model, PubMedBERT(abstracts + full text), PubMedBERT(only abstracts), Unified Medical Language System (UMLS) BERT, Bio BERT, and Bio-clinical BERT. We performed two case studies: physical activity and excessive diet, in order to validate the effectiveness of BERT models in classifying lifestyle factors for AD. These models were compared on the developed Gold Standard Corpus (GSC) on the two case studies. The PubmedBERT(Abs) model achieved the best performance for physical activity, with its precision, recall, and F-1 scores of 0.96, 0.96, and 0.96, respectively. Regarding classifying excessive diet, the Bio BERT model showed the highest performance with perfect precision, recall, and F-1 scores. The proposed approach leveraging weak supervision could significantly increase the sample size, which is required for training the deep learning models. The study also demonstrates the effectiveness of BERT models for extracting lifestyle factors for Alzheimer's disease from clinical notes.
Social determinants of health in the era of artificial intelligence with electronic health records: A systematic review
Jan 22, 2021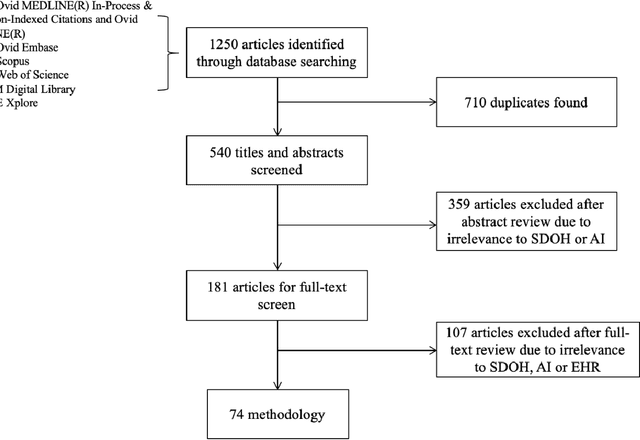
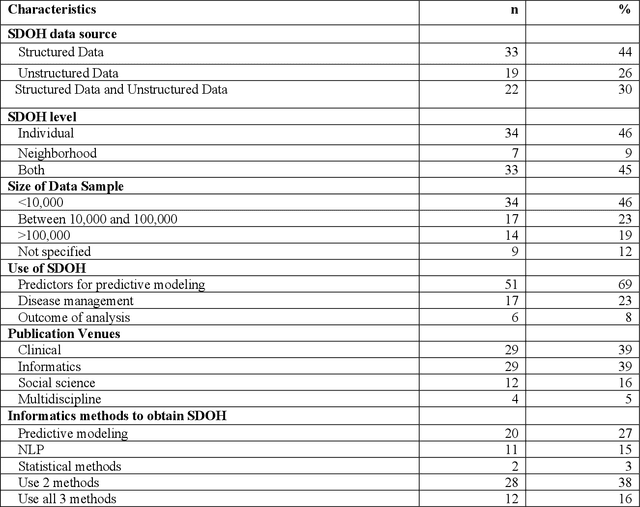
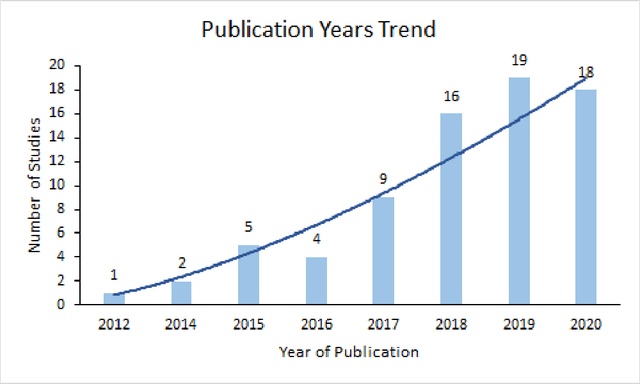
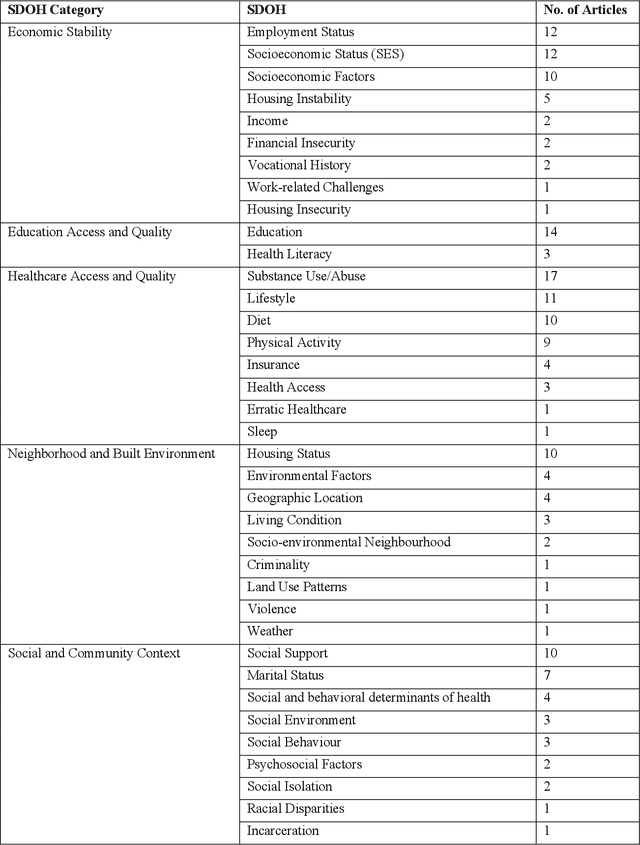
Abstract:There is growing evidence showing the significant role of social determinant of health (SDOH) on a wide variety of health outcomes. In the era of artificial intelligence (AI), electronic health records (EHRs) have been widely used to conduct observational studies. However, how to make the best of SDOH information from EHRs is yet to be studied. In this paper, we systematically reviewed recently published papers and provided a methodology review of AI methods using the SDOH information in EHR data. A total of 1250 articles were retrieved from the literature between 2010 and 2020, and 74 papers were included in this review after abstract and full-text screening. We summarized these papers in terms of general characteristics (including publication years, venues, countries etc.), SDOH types, disease areas, study outcomes, AI methods to extract SDOH from EHRs and AI methods using SDOH for healthcare outcomes. Finally, we conclude this paper with discussion on the current trends, challenges, and future directions on using SDOH from EHRs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge