Fang Yu
scGHSOM: Hierarchical clustering and visualization of single-cell and CRISPR data using growing hierarchical SOM
Jul 24, 2024Abstract:High-dimensional single-cell data poses significant challenges in identifying underlying biological patterns due to the complexity and heterogeneity of cellular states. We propose a comprehensive gene-cell dependency visualization via unsupervised clustering, Growing Hierarchical Self-Organizing Map (GHSOM), specifically designed for analyzing high-dimensional single-cell data like single-cell sequencing and CRISPR screens. GHSOM is applied to cluster samples in a hierarchical structure such that the self-growth structure of clusters satisfies the required variations between and within. We propose a novel Significant Attributes Identification Algorithm to identify features that distinguish clusters. This algorithm pinpoints attributes with minimal variation within a cluster but substantial variation between clusters. These key attributes can then be used for targeted data retrieval and downstream analysis. Furthermore, we present two innovative visualization tools: Cluster Feature Map and Cluster Distribution Map. The Cluster Feature Map highlights the distribution of specific features across the hierarchical structure of GHSOM clusters. This allows for rapid visual assessment of cluster uniqueness based on chosen features. The Cluster Distribution Map depicts leaf clusters as circles on the GHSOM grid, with circle size reflecting cluster data size and color customizable to visualize features like cell type or other attributes. We apply our analysis to three single-cell datasets and one CRISPR dataset (cell-gene database) and evaluate clustering methods with internal and external CH and ARI scores. GHSOM performs well, being the best performer in internal evaluation (CH=4.2). In external evaluation, GHSOM has the third-best performance of all methods.
Automated Hit-frame Detection for Badminton Match Analysis
Aug 02, 2023



Abstract:Sports professionals constantly under pressure to perform at the highest level can benefit from sports analysis, which allows coaches and players to reduce manual efforts and systematically evaluate their performance using automated tools. This research aims to advance sports analysis in badminton, systematically detecting hit-frames automatically from match videos using modern deep learning techniques. The data included in hit-frames can subsequently be utilized to synthesize players' strokes and on-court movement, as well as for other downstream applications such as analyzing training tasks and competition strategy. The proposed approach in this study comprises several automated procedures like rally-wise video trimming, player and court keypoints detection, shuttlecock flying direction prediction, and hit-frame detection. In the study, we achieved 99% accuracy on shot angle recognition for video trimming, over 92% accuracy for applying player keypoints sequences on shuttlecock flying direction prediction, and reported the evaluation results of rally-wise video trimming and hit-frame detection.
Extracting Lifestyle Factors for Alzheimer's Disease from Clinical Notes Using Deep Learning with Weak Supervision
Jan 25, 2021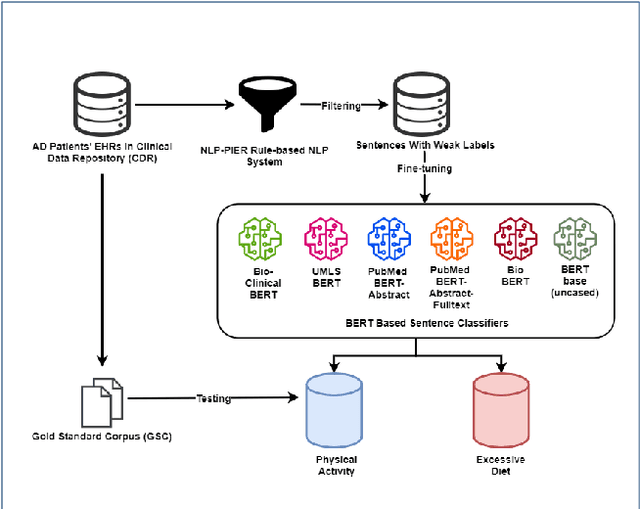
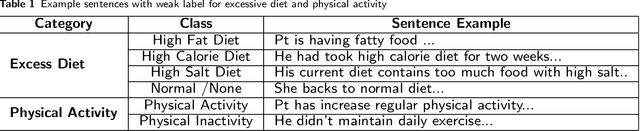
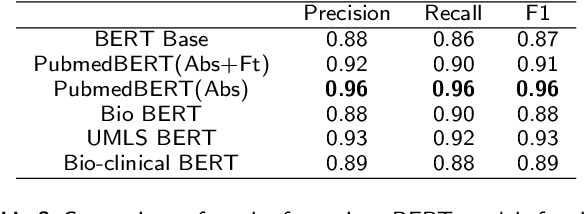
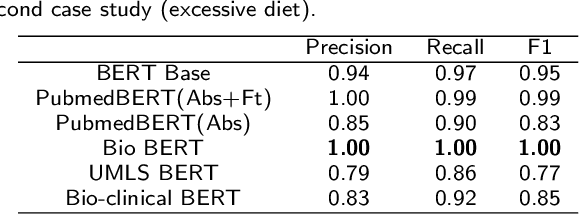
Abstract:Since no effective therapies exist for Alzheimer's disease (AD), prevention has become more critical through lifestyle factor changes and interventions. Analyzing electronic health records (EHR) of patients with AD can help us better understand lifestyle's effect on AD. However, lifestyle information is typically stored in clinical narratives. Thus, the objective of the study was to demonstrate the feasibility of natural language processing (NLP) models to classify lifestyle factors (e.g., physical activity and excessive diet) from clinical texts. We automatically generated labels for the training data by using a rule-based NLP algorithm. We conducted weak supervision for pre-trained Bidirectional Encoder Representations from Transformers (BERT) models on the weakly labeled training corpus. These models include the BERT base model, PubMedBERT(abstracts + full text), PubMedBERT(only abstracts), Unified Medical Language System (UMLS) BERT, Bio BERT, and Bio-clinical BERT. We performed two case studies: physical activity and excessive diet, in order to validate the effectiveness of BERT models in classifying lifestyle factors for AD. These models were compared on the developed Gold Standard Corpus (GSC) on the two case studies. The PubmedBERT(Abs) model achieved the best performance for physical activity, with its precision, recall, and F-1 scores of 0.96, 0.96, and 0.96, respectively. Regarding classifying excessive diet, the Bio BERT model showed the highest performance with perfect precision, recall, and F-1 scores. The proposed approach leveraging weak supervision could significantly increase the sample size, which is required for training the deep learning models. The study also demonstrates the effectiveness of BERT models for extracting lifestyle factors for Alzheimer's disease from clinical notes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge