Pierre Jannin
DSTED: Decoupling Temporal Stabilization and Discriminative Enhancement for Surgical Workflow Recognition
Dec 22, 2025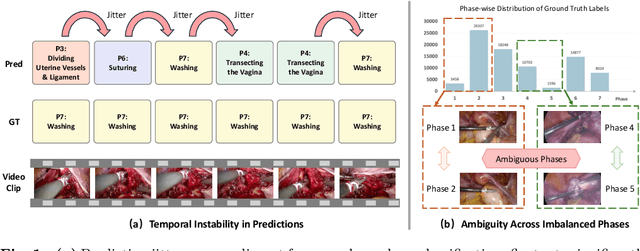
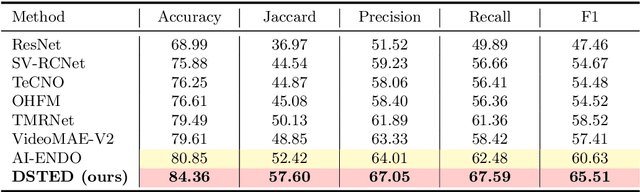
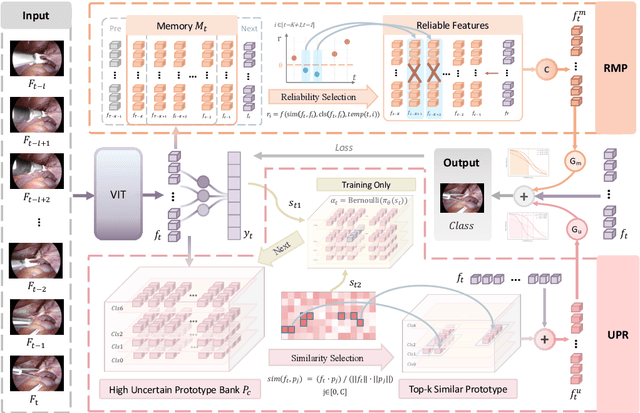
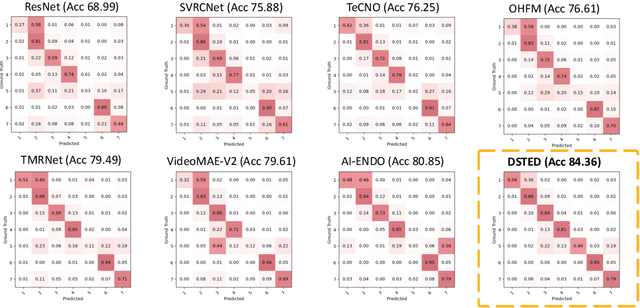
Abstract:Purpose: Surgical workflow recognition enables context-aware assistance and skill assessment in computer-assisted interventions. Despite recent advances, current methods suffer from two critical challenges: prediction jitter across consecutive frames and poor discrimination of ambiguous phases. This paper aims to develop a stable framework by selectively propagating reliable historical information and explicitly modeling uncertainty for hard sample enhancement. Methods: We propose a dual-pathway framework DSTED with Reliable Memory Propagation (RMP) and Uncertainty-Aware Prototype Retrieval (UPR). RMP maintains temporal coherence by filtering and fusing high-confidence historical features through multi-criteria reliability assessment. UPR constructs learnable class-specific prototypes from high-uncertainty samples and performs adaptive prototype matching to refine ambiguous frame representations. Finally, a confidence-driven gate dynamically balances both pathways based on prediction certainty. Results: Our method achieves state-of-the-art performance on AutoLaparo-hysterectomy with 84.36% accuracy and 65.51% F1-score, surpassing the second-best method by 3.51% and 4.88% respectively. Ablations reveal complementary gains from RMP (2.19%) and UPR (1.93%), with synergistic effects when combined. Extensive analysis confirms substantial reduction in temporal jitter and marked improvement on challenging phase transitions. Conclusion: Our dual-pathway design introduces a novel paradigm for stable workflow recognition, demonstrating that decoupling the modeling of temporal consistency and phase ambiguity yields superior performance and clinical applicability.
Measuring proximity to standard planes during fetal brain ultrasound scanning
Apr 10, 2024



Abstract:This paper introduces a novel pipeline designed to bring ultrasound (US) plane pose estimation closer to clinical use for more effective navigation to the standard planes (SPs) in the fetal brain. We propose a semi-supervised segmentation model utilizing both labeled SPs and unlabeled 3D US volume slices. Our model enables reliable segmentation across a diverse set of fetal brain images. Furthermore, the model incorporates a classification mechanism to identify the fetal brain precisely. Our model not only filters out frames lacking the brain but also generates masks for those containing it, enhancing the relevance of plane pose regression in clinical settings. We focus on fetal brain navigation from 2D ultrasound (US) video analysis and combine this model with a US plane pose regression network to provide sensorless proximity detection to SPs and non-SPs planes; we emphasize the importance of proximity detection to SPs for guiding sonographers, offering a substantial advantage over traditional methods by allowing earlier and more precise adjustments during scanning. We demonstrate the practical applicability of our approach through validation on real fetal scan videos obtained from sonographers of varying expertise levels. Our findings demonstrate the potential of our approach to complement existing fetal US technologies and advance prenatal diagnostic practices.
Why is the winner the best?
Mar 30, 2023
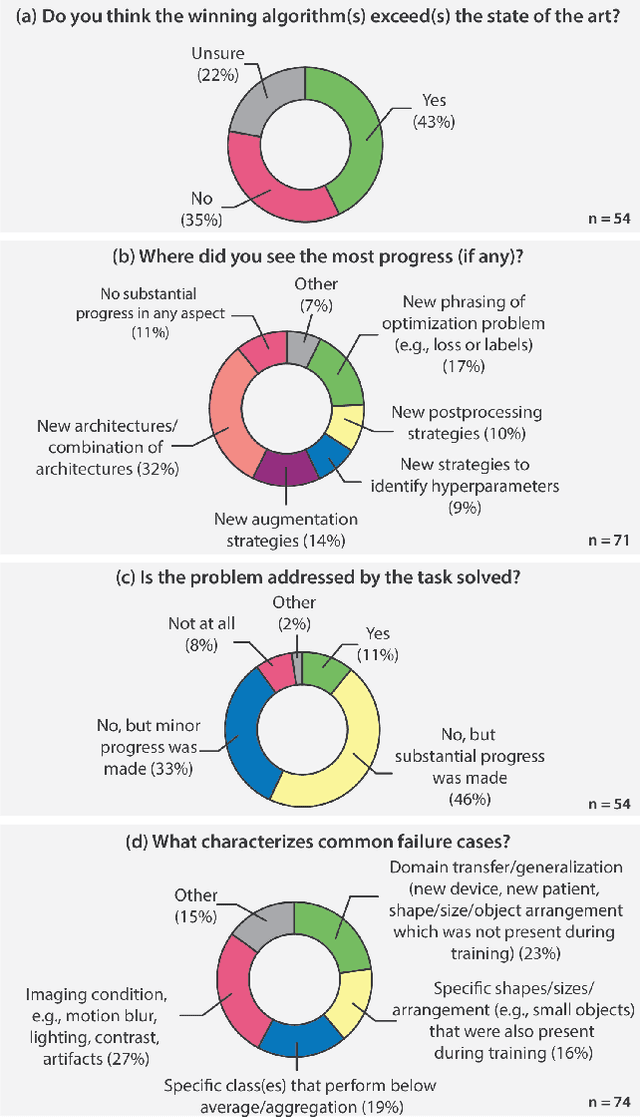
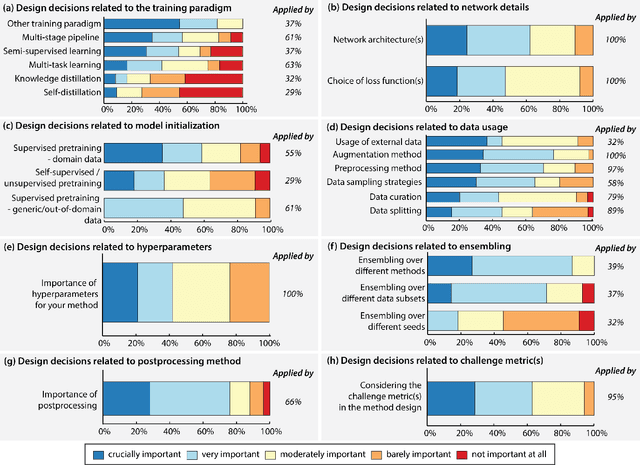
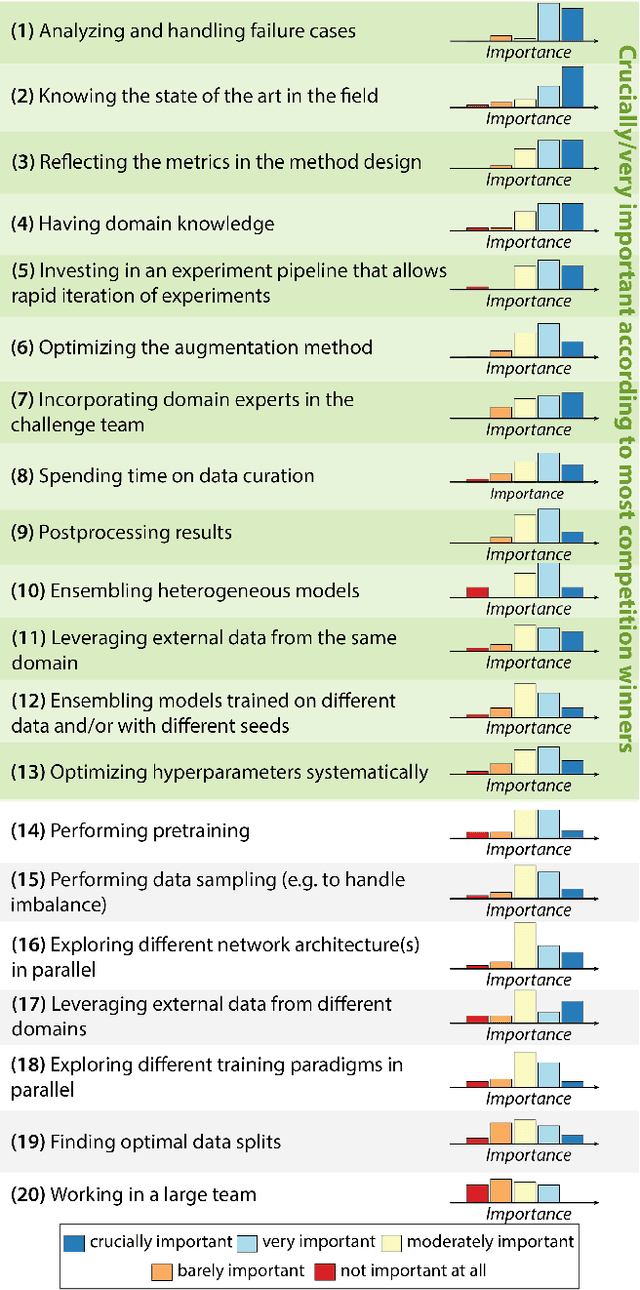
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022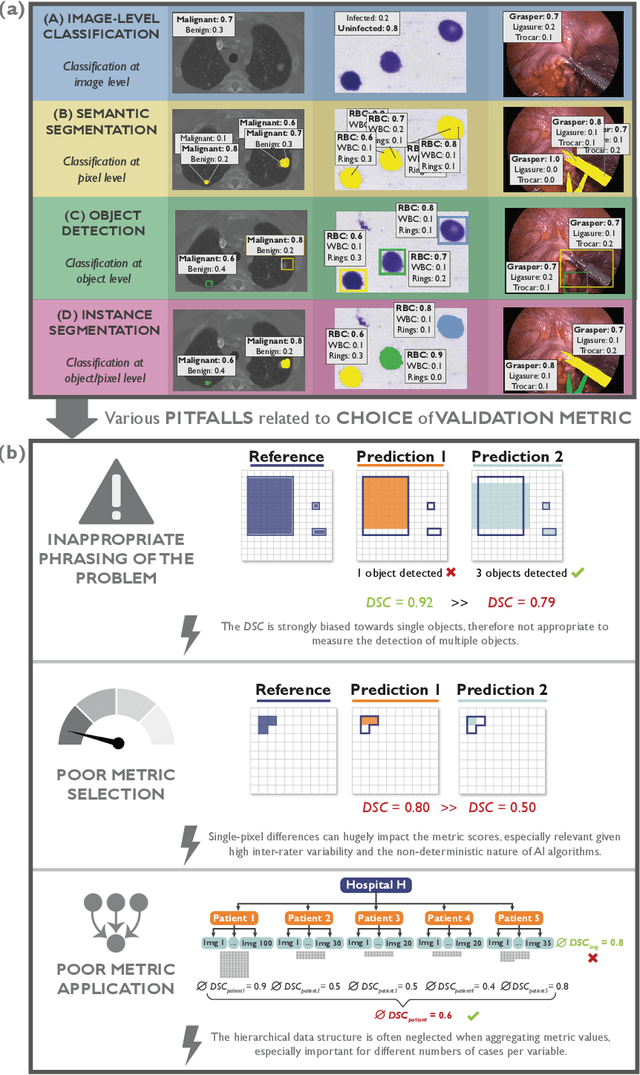
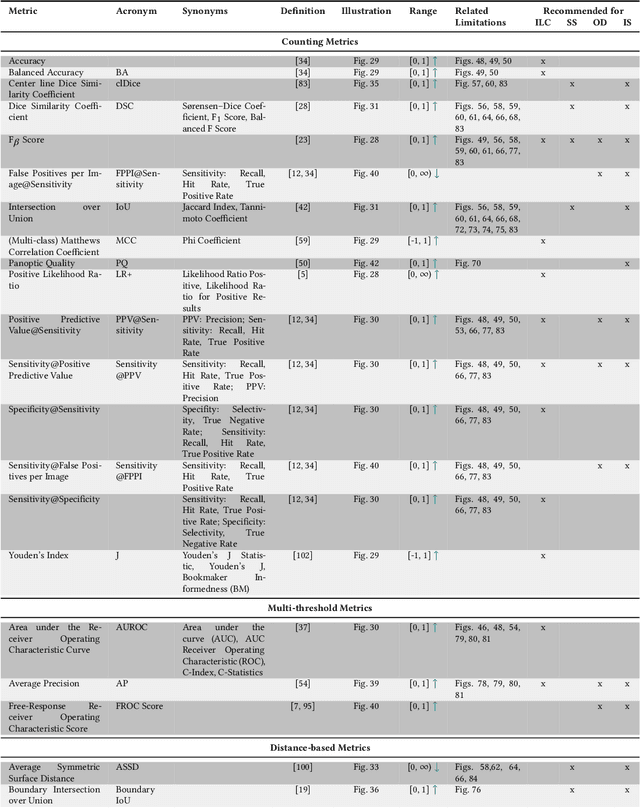
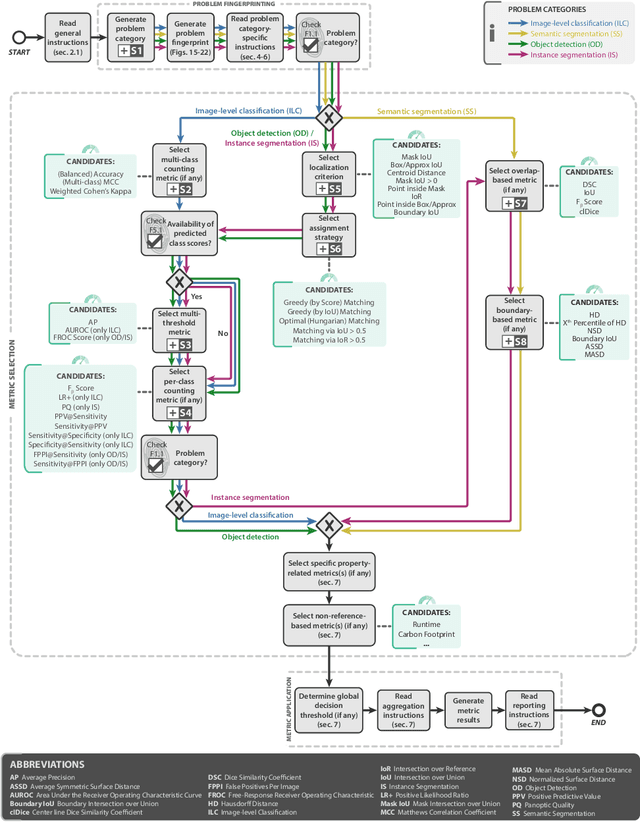
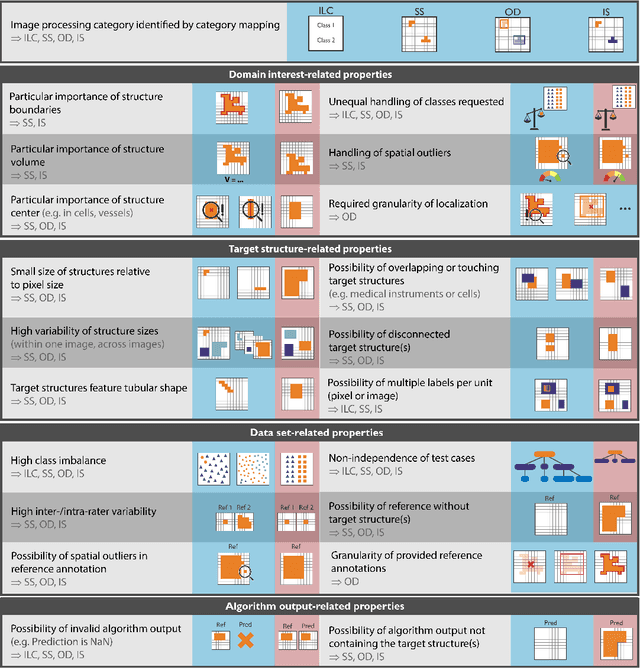
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
PEg TRAnsfer Workflow recognition challenge report: Does multi-modal data improve recognition?
Feb 11, 2022
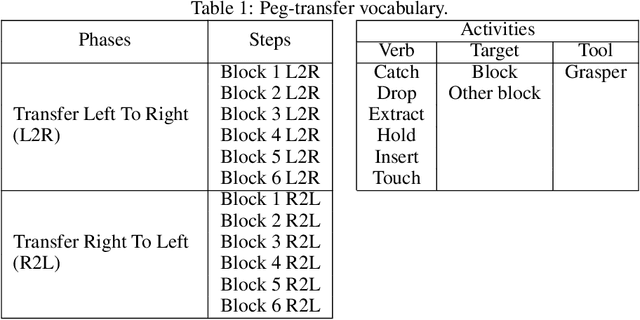
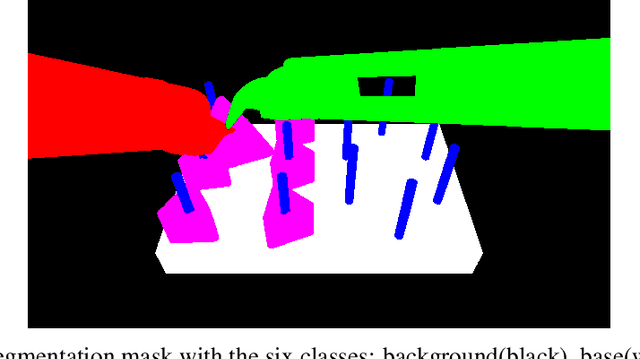
Abstract:This paper presents the design and results of the "PEg TRAnsfert Workflow recognition" (PETRAW) challenge whose objective was to develop surgical workflow recognition methods based on one or several modalities, among video, kinematic, and segmentation data, in order to study their added value. The PETRAW challenge provided a data set of 150 peg transfer sequences performed on a virtual simulator. This data set was composed of videos, kinematics, semantic segmentation, and workflow annotations which described the sequences at three different granularity levels: phase, step, and activity. Five tasks were proposed to the participants: three of them were related to the recognition of all granularities with one of the available modalities, while the others addressed the recognition with a combination of modalities. Average application-dependent balanced accuracy (AD-Accuracy) was used as evaluation metric to take unbalanced classes into account and because it is more clinically relevant than a frame-by-frame score. Seven teams participated in at least one task and four of them in all tasks. Best results are obtained with the use of the video and the kinematics data with an AD-Accuracy between 93% and 90% for the four teams who participated in all tasks. The improvement between video/kinematic-based methods and the uni-modality ones was significant for all of the teams. However, the difference in testing execution time between the video/kinematic-based and the kinematic-based methods has to be taken into consideration. Is it relevant to spend 20 to 200 times more computing time for less than 3% of improvement? The PETRAW data set is publicly available at www.synapse.org/PETRAW to encourage further research in surgical workflow recognition.
Common Limitations of Image Processing Metrics: A Picture Story
Apr 13, 2021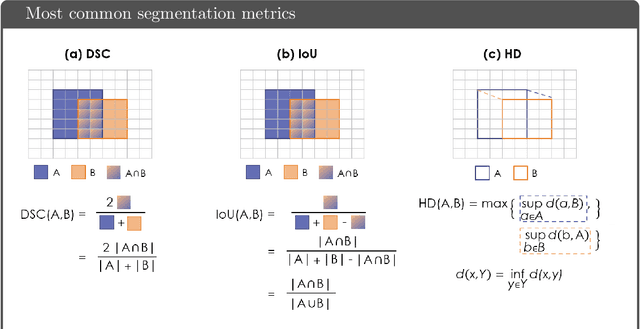
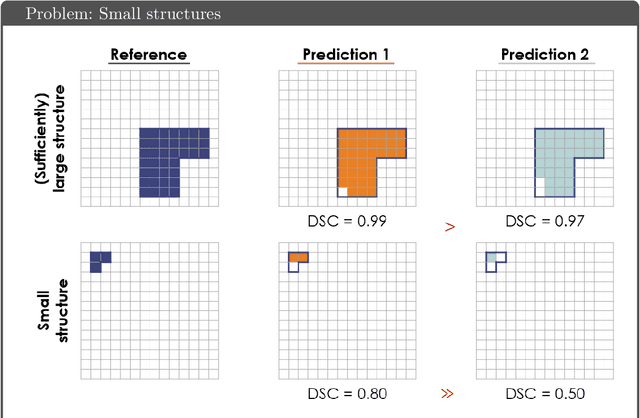
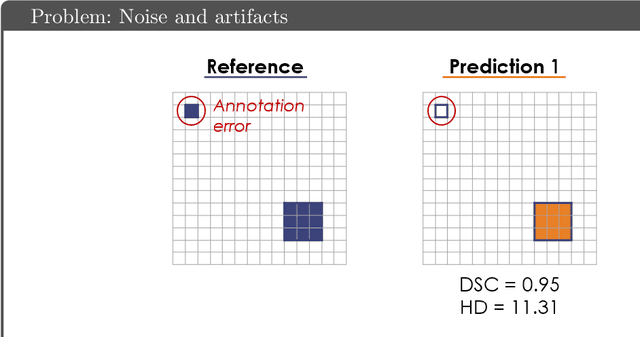
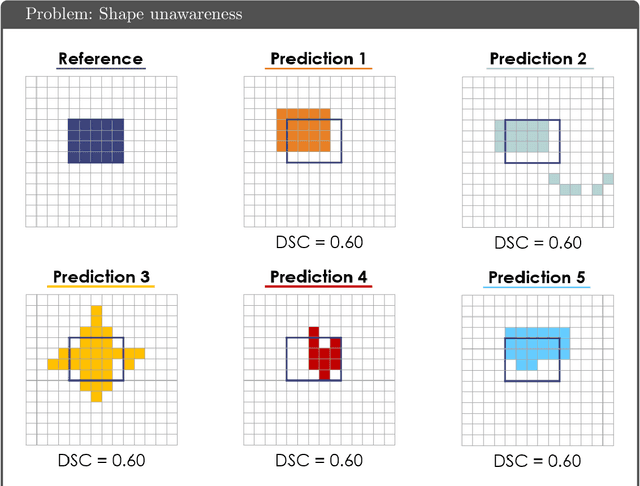
Abstract:While the importance of automatic image analysis is increasing at an enormous pace, recent meta-research revealed major flaws with respect to algorithm validation. Specifically, performance metrics are key for objective, transparent and comparative performance assessment, but relatively little attention has been given to the practical pitfalls when using specific metrics for a given image analysis task. A common mission of several international initiatives is therefore to provide researchers with guidelines and tools to choose the performance metrics in a problem-aware manner. This dynamically updated document has the purpose to illustrate important limitations of performance metrics commonly applied in the field of image analysis. The current version is based on a Delphi process on metrics conducted by an international consortium of image analysis experts.
MIcro-Surgical Anastomose Workflow recognition challenge report
Mar 24, 2021



Abstract:The "MIcro-Surgical Anastomose Workflow recognition on training sessions" (MISAW) challenge provided a data set of 27 sequences of micro-surgical anastomosis on artificial blood vessels. This data set was composed of videos, kinematics, and workflow annotations described at three different granularity levels: phase, step, and activity. The participants were given the option to use kinematic data and videos to develop workflow recognition models. Four tasks were proposed to the participants: three of them were related to the recognition of surgical workflow at three different granularity levels, while the last one addressed the recognition of all granularity levels in the same model. One ranking was made for each task. We used the average application-dependent balanced accuracy (AD-Accuracy) as the evaluation metric. This takes unbalanced classes into account and it is more clinically relevant than a frame-by-frame score. Six teams, including a non-competing team, participated in at least one task. All models employed deep learning models, such as CNN or RNN. The best models achieved more than 95% AD-Accuracy for phase recognition, 80% for step recognition, 60% for activity recognition, and 75% for all granularity levels. For high levels of granularity (i.e., phases and steps), the best models had a recognition rate that may be sufficient for applications such as prediction of remaining surgical time or resource management. However, for activities, the recognition rate was still low for applications that can be employed clinically. The MISAW data set is publicly available to encourage further research in surgical workflow recognition. It can be found at www.synapse.org/MISAW
Surgical Data Science -- from Concepts to Clinical Translation
Oct 30, 2020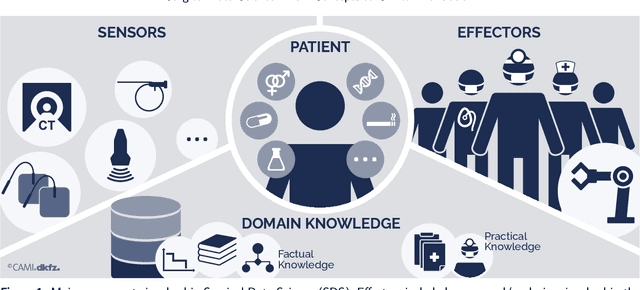
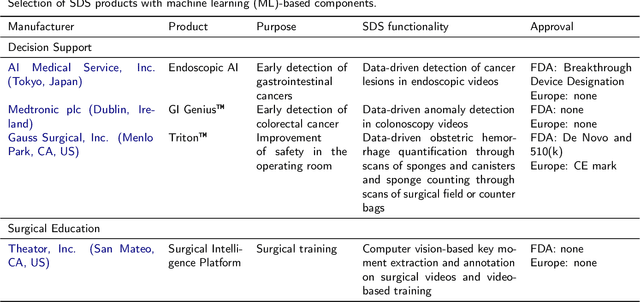
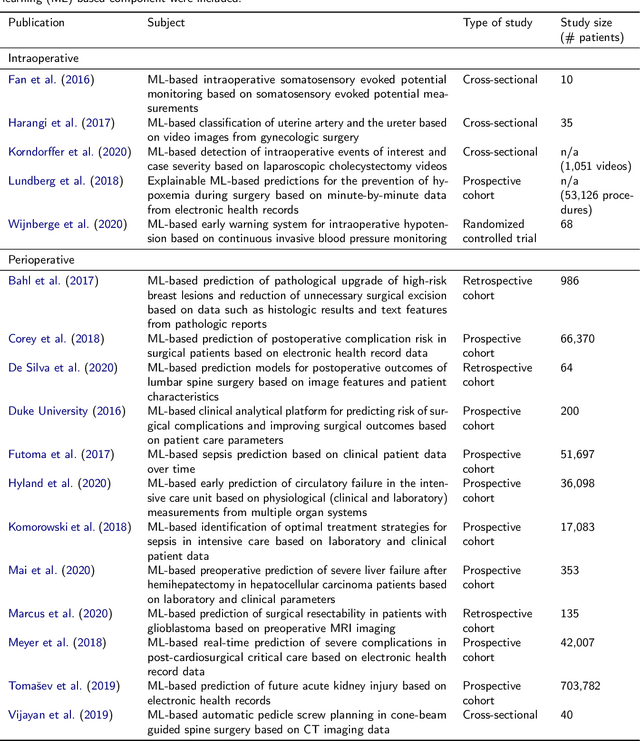
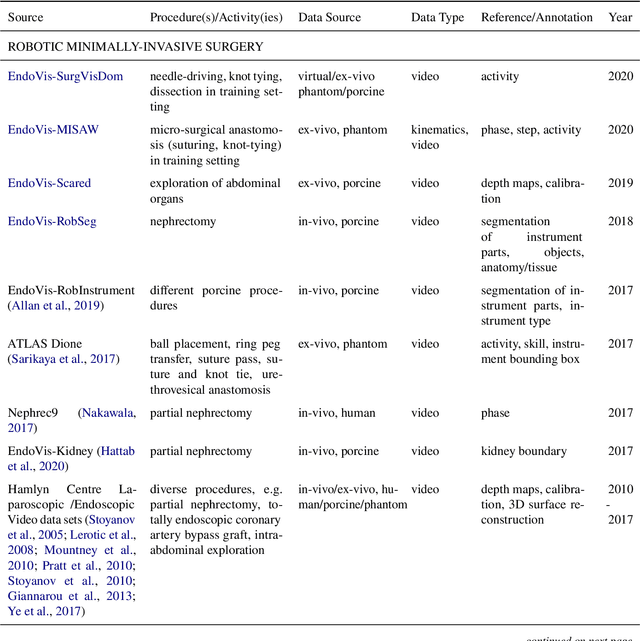
Abstract:Recent developments in data science in general and machine learning in particular have transformed the way experts envision the future of surgery. Surgical data science is a new research field that aims to improve the quality of interventional healthcare through the capture, organization, analysis and modeling of data. While an increasing number of data-driven approaches and clinical applications have been studied in the fields of radiological and clinical data science, translational success stories are still lacking in surgery. In this publication, we shed light on the underlying reasons and provide a roadmap for future advances in the field. Based on an international workshop involving leading researchers in the field of surgical data science, we review current practice, key achievements and initiatives as well as available standards and tools for a number of topics relevant to the field, namely (1) technical infrastructure for data acquisition, storage and access in the presence of regulatory constraints, (2) data annotation and sharing and (3) data analytics. Drawing from this extensive review, we present current challenges for technology development and (4) describe a roadmap for faster clinical translation and exploitation of the full potential of surgical data science.
Towards Generalizable Surgical Activity Recognition Using Spatial Temporal Graph Convolutional Networks
Jan 16, 2020



Abstract:Purpose Modeling and recognition of surgical activities poses an interesting research problem. Although a number of recent works studied automatic recognition of surgical activities, generalizability of these works across different tasks and different datasets remains a challenge. We introduce a modality that is robust to scene variation, based on spatial temporal graph representations of surgical tools in videos for surgical activity recognition. Methods To show its effectiveness, we model and recognize surgical gestures with the proposed modality. We construct spatial graphs connecting the joint pose estimations of surgical tools. Then, we connect each joint to the corresponding joint in the consecutive frames forming inter-frame edges representing the trajectory of the joint over time. We then learn hierarchical spatial temporal graph representations using Spatial Temporal Graph Convolutional Networks (ST-GCN). Results Our experimental results show that learned spatial temporal graph representations of surgical videos perform well in surgical gesture recognition even when used individually. We experiment with the Suturing task of the JIGSAWS dataset where the chance baseline for gesture recognition is 10%. Our results demonstrate 68% average accuracy which suggests a significant improvement. Conclusions Our experimental results show that our model learns meaningful representations.These learned representations can be used either individually, in cascades or as a complementary modality in surgical activity recognition, therefore provide a benchmark. To our knowledge, our paper is the first to use spatial temporal graph representations based on pose estimations of surgical tools in surgical activity recognition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge