Mikael Brudfors
Automated Surgical Skill Assessment in Endoscopic Pituitary Surgery using Real-time Instrument Tracking on a High-fidelity Bench-top Phantom
Sep 25, 2024Abstract:Improved surgical skill is generally associated with improved patient outcomes, although assessment is subjective; labour-intensive; and requires domain specific expertise. Automated data driven metrics can alleviate these difficulties, as demonstrated by existing machine learning instrument tracking models in minimally invasive surgery. However, these models have been tested on limited datasets of laparoscopic surgery, with a focus on isolated tasks and robotic surgery. In this paper, a new public dataset is introduced, focusing on simulated surgery, using the nasal phase of endoscopic pituitary surgery as an exemplar. Simulated surgery allows for a realistic yet repeatable environment, meaning the insights gained from automated assessment can be used by novice surgeons to hone their skills on the simulator before moving to real surgery. PRINTNet (Pituitary Real-time INstrument Tracking Network) has been created as a baseline model for this automated assessment. Consisting of DeepLabV3 for classification and segmentation; StrongSORT for tracking; and the NVIDIA Holoscan SDK for real-time performance, PRINTNet achieved 71.9% Multiple Object Tracking Precision running at 22 Frames Per Second. Using this tracking output, a Multilayer Perceptron achieved 87% accuracy in predicting surgical skill level (novice or expert), with the "ratio of total procedure time to instrument visible time" correlated with higher surgical skill. This therefore demonstrates the feasibility of automated surgical skill assessment in simulated endoscopic pituitary surgery. The new publicly available dataset can be found here: https://doi.org/10.5522/04/26511049.
Large-kernel Attention for Efficient and Robust Brain Lesion Segmentation
Aug 14, 2023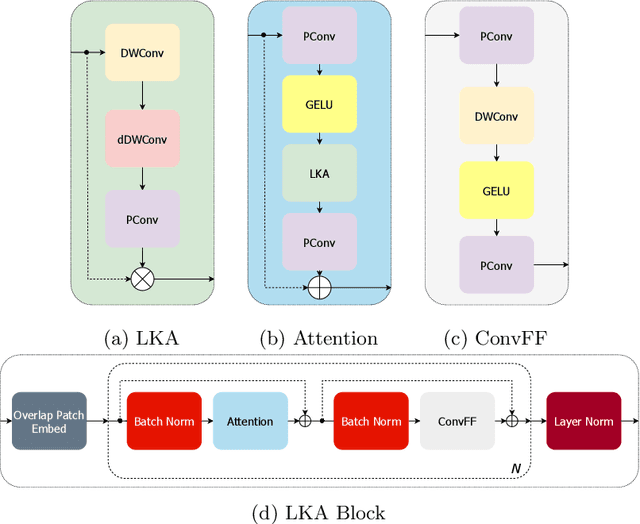
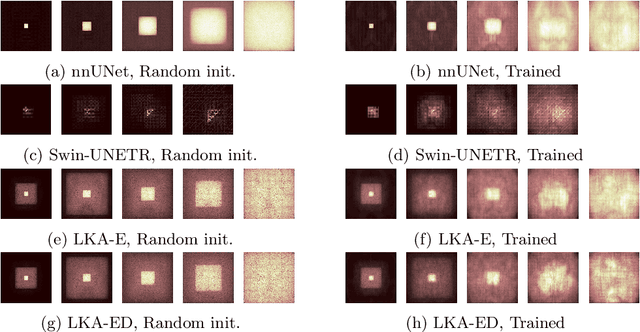
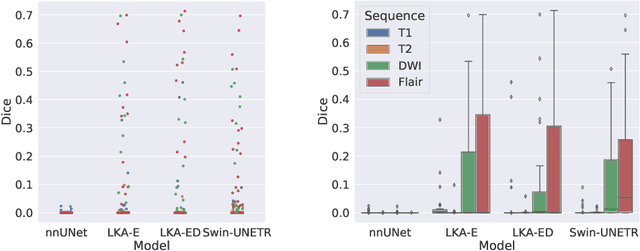
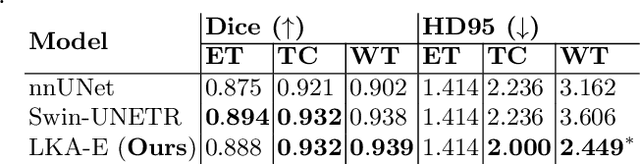
Abstract:Vision transformers are effective deep learning models for vision tasks, including medical image segmentation. However, they lack efficiency and translational invariance, unlike convolutional neural networks (CNNs). To model long-range interactions in 3D brain lesion segmentation, we propose an all-convolutional transformer block variant of the U-Net architecture. We demonstrate that our model provides the greatest compromise in three factors: performance competitive with the state-of-the-art; parameter efficiency of a CNN; and the favourable inductive biases of a transformer. Our public implementation is available at https://github.com/liamchalcroft/MDUNet .
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Fitting Segmentation Networks on Varying Image Resolutions using Splatting
Jun 15, 2022


Abstract:Data used in image segmentation are not always defined on the same grid. This is particularly true for medical images, where the resolution, field-of-view and orientation can differ across channels and subjects. Images and labels are therefore commonly resampled onto the same grid, as a pre-processing step. However, the resampling operation introduces partial volume effects and blurring, thereby changing the effective resolution and reducing the contrast between structures. In this paper we propose a splat layer, which automatically handles resolution mismatches in the input data. This layer pushes each image onto a mean space where the forward pass is performed. As the splat operator is the adjoint to the resampling operator, the mean-space prediction can be pulled back to the native label space, where the loss function is computed. Thus, the need for explicit resolution adjustment using interpolation is removed. We show on two publicly available datasets, with simulated and real multi-modal magnetic resonance images, that this model improves segmentation results compared to resampling as a pre-processing step.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021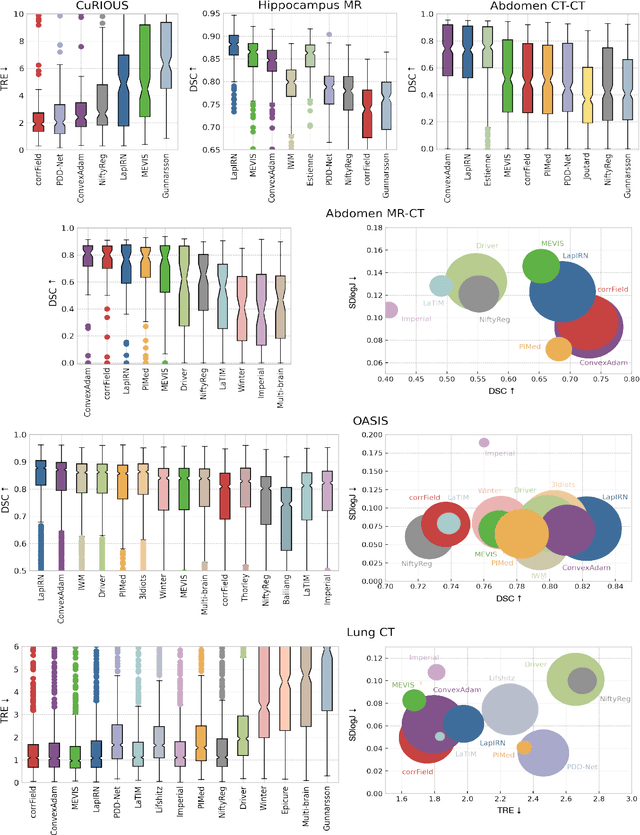
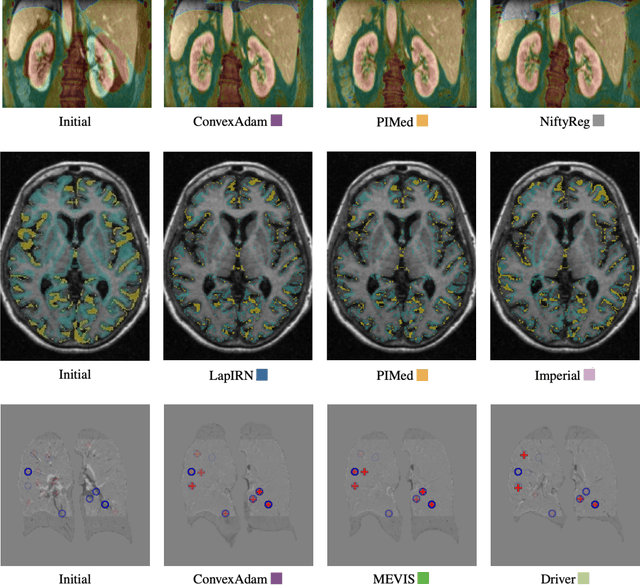
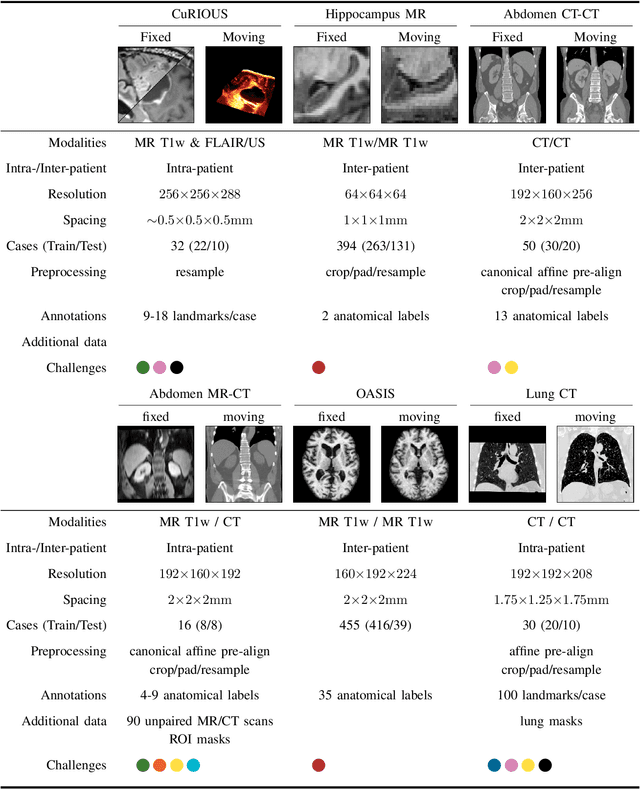
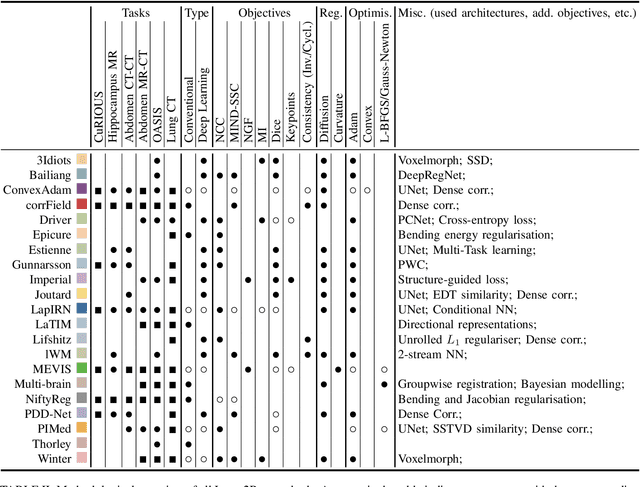
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
Factorisation-based Image Labelling
Nov 19, 2021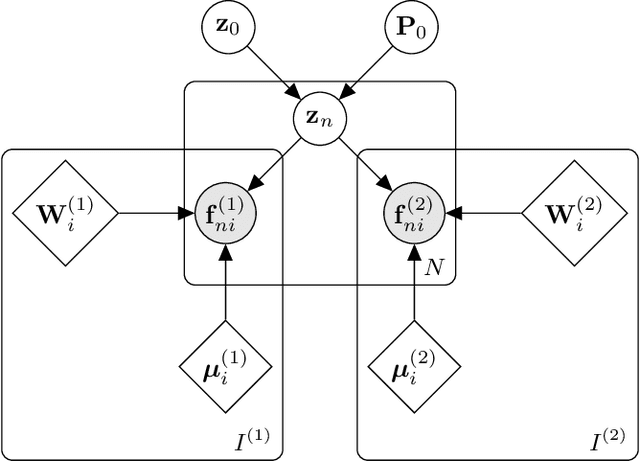
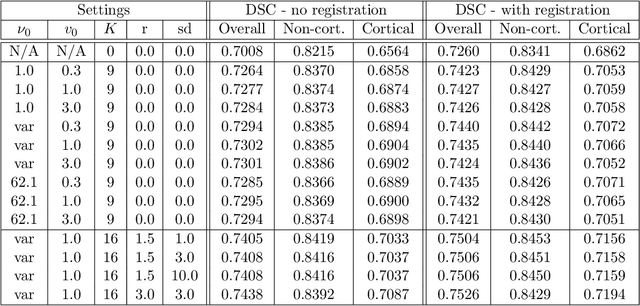
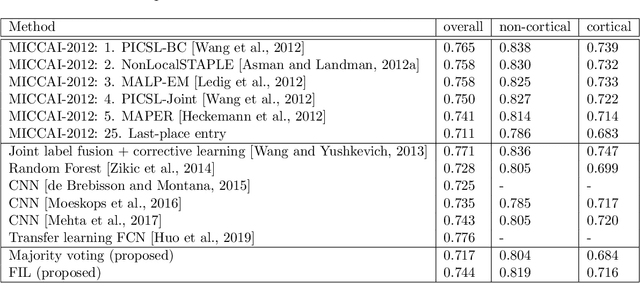
Abstract:Segmentation of brain magnetic resonance images (MRI) into anatomical regions is a useful task in neuroimaging. Manual annotation is time consuming and expensive, so having a fully automated and general purpose brain segmentation algorithm is highly desirable. To this end, we propose a patched-based label propagation approach based on a generative model with latent variables. Once trained, our Factorisation-based Image Labelling (FIL) model is able to label target images with a variety of image contrasts. We compare the effectiveness of our proposed model against the state-of-the-art using data from the MICCAI 2012 Grand Challenge and Workshop on Multi-Atlas Labeling. As our approach is intended to be general purpose, we also assess how well it can handle domain shift by labelling images of the same subjects acquired with different MR contrasts.
Correcting inter-scan motion artefacts in quantitative R1 mapping at 7T
Aug 24, 2021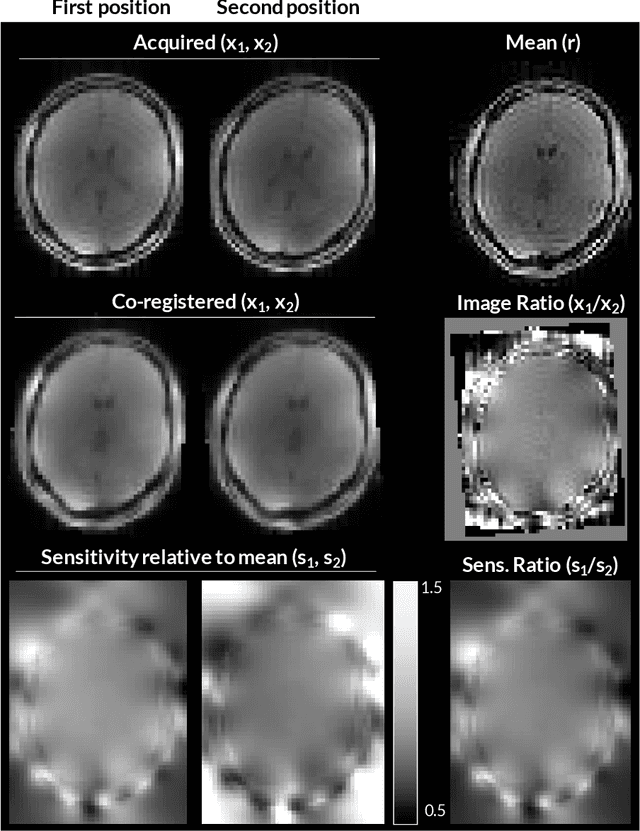
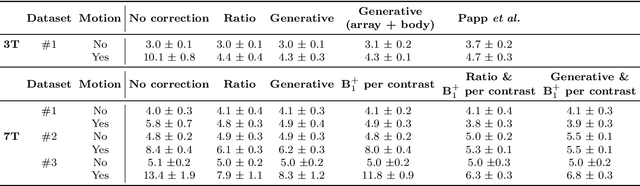
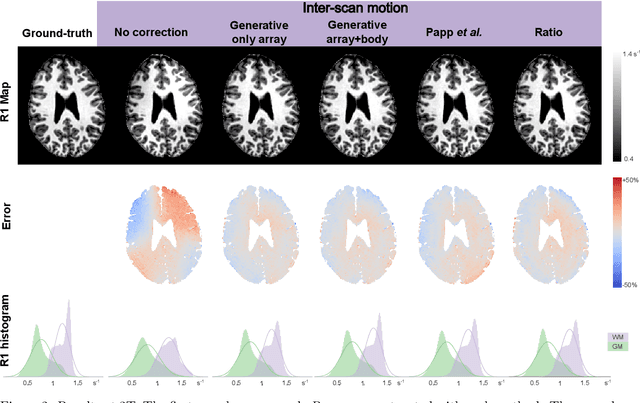
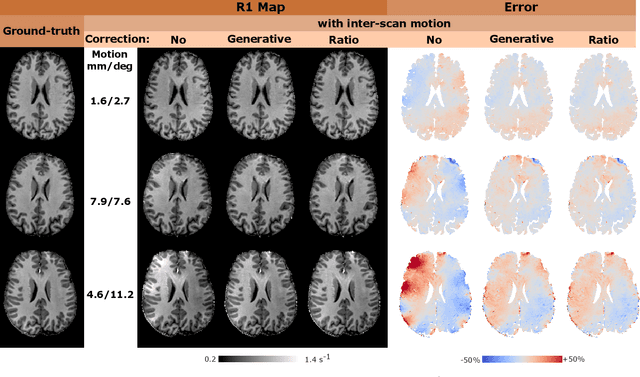
Abstract:Purpose: Inter-scan motion is a substantial source of error in $R_1$ estimation, and can be expected to increase at 7T where $B_1$ fields are more inhomogeneous. The established correction scheme does not translate to 7T since it requires a body coil reference. Here we introduce two alternatives that outperform the established method. Since they compute relative sensitivities they do not require body coil images. Theory: The proposed methods use coil-combined magnitude images to obtain the relative coil sensitivities. The first method efficiently computes the relative sensitivities via a simple ratio; the second by fitting a more sophisticated generative model. Methods: $R_1$ maps were computed using the variable flip angle (VFA) approach. Multiple datasets were acquired at 3T and 7T, with and without motion between the acquisition of the VFA volumes. $R_1$ maps were constructed without correction, with the proposed corrections, and (at 3T) with the previously established correction scheme. Results: At 3T, the proposed methods outperform the baseline method. Inter-scan motion artefacts were also reduced at 7T. However, reproducibility only converged on that of the no motion condition if position-specific transmit field effects were also incorporated. Conclusion: The proposed methods simplify inter-scan motion correction of $R_1$ maps and are applicable at both 3T and 7T, where a body coil is typically not available. The open-source code for all methods is made publicly available.
An MRF-UNet Product of Experts for Image Segmentation
Apr 12, 2021



Abstract:While convolutional neural networks (CNNs) trained by back-propagation have seen unprecedented success at semantic segmentation tasks, they are known to struggle on out-of-distribution data. Markov random fields (MRFs) on the other hand, encode simpler distributions over labels that, although less flexible than UNets, are less prone to over-fitting. In this paper, we propose to fuse both strategies by computing the product of distributions of a UNet and an MRF. As this product is intractable, we solve for an approximate distribution using an iterative mean-field approach. The resulting MRF-UNet is trained jointly by back-propagation. Compared to other works using conditional random fields (CRFs), the MRF has no dependency on the imaging data, which should allow for less over-fitting. We show on 3D neuroimaging data that this novel network improves generalisation to out-of-distribution samples. Furthermore, it allows the overall number of parameters to be reduced while preserving high accuracy. These results suggest that a classic MRF smoothness prior can allow for less over-fitting when principally integrated into a CNN model. Our implementation is available at https://github.com/balbasty/nitorch.
Model-based multi-parameter mapping
Feb 02, 2021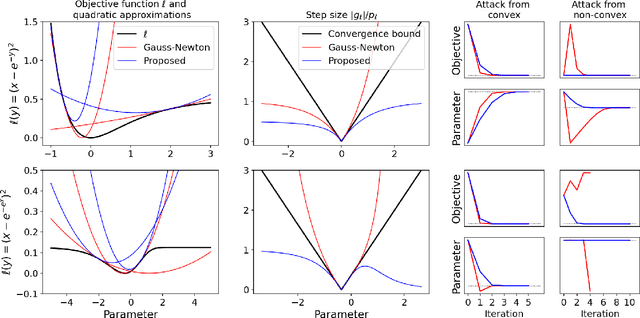
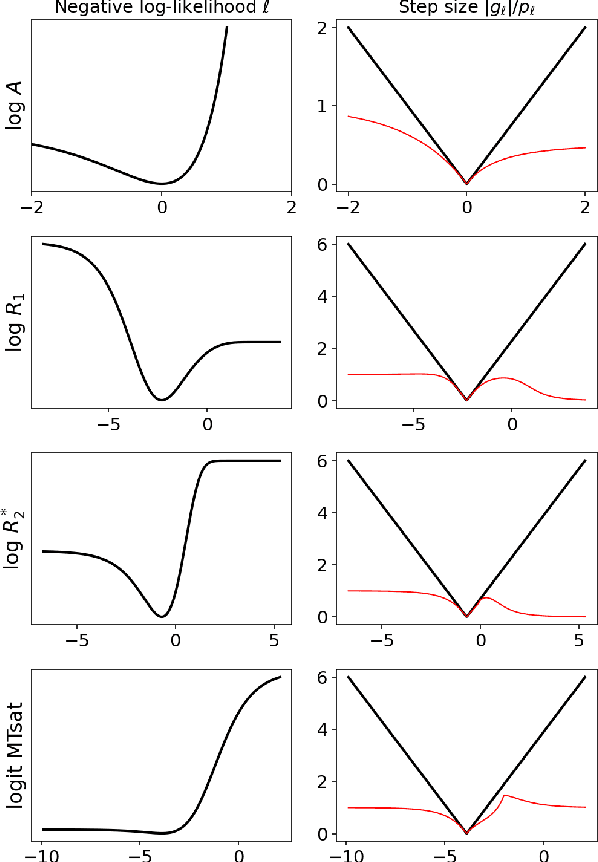
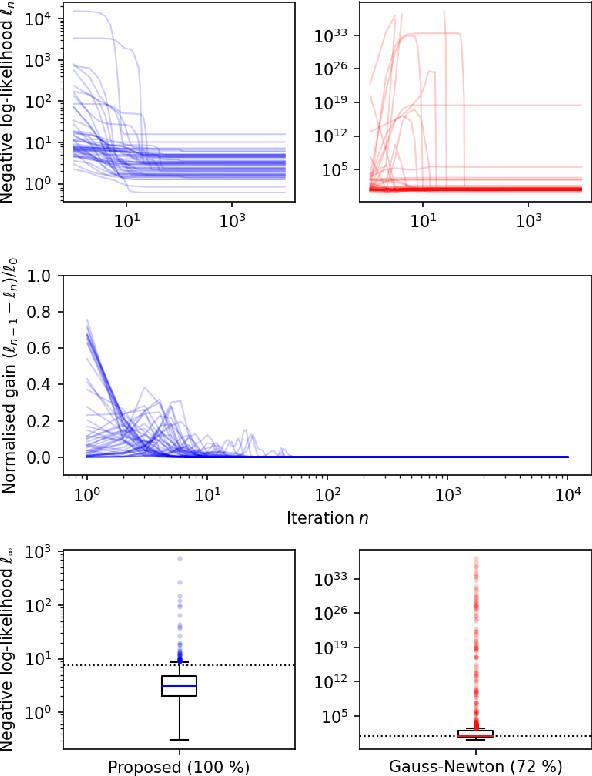
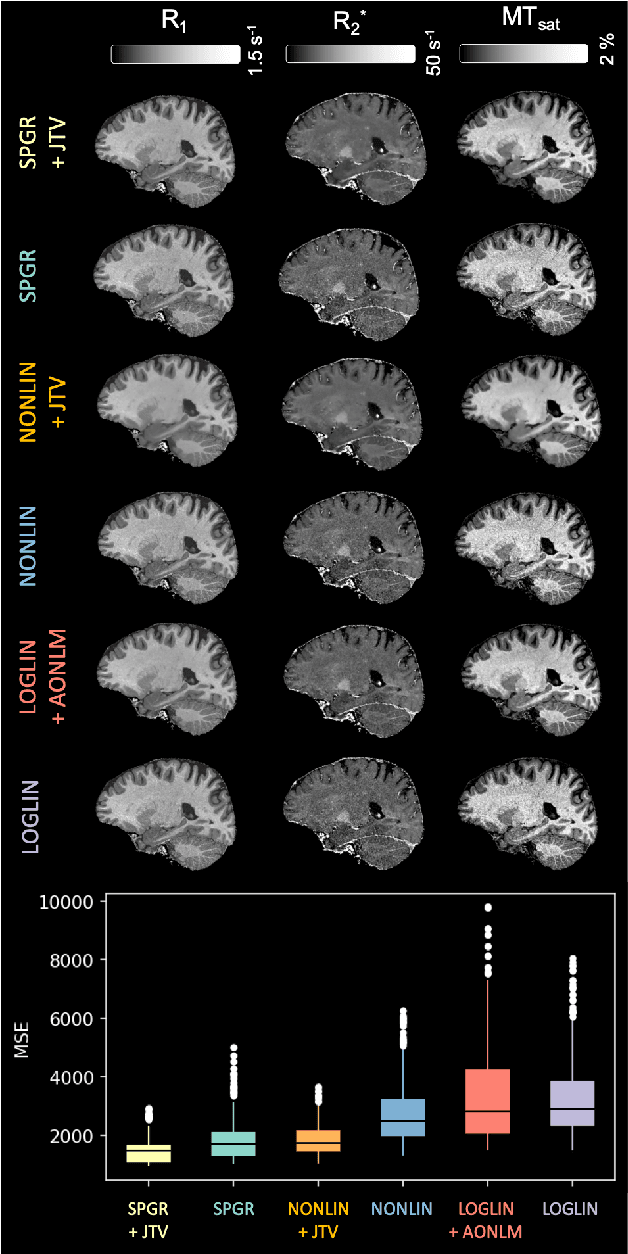
Abstract:Quantitative MR imaging is increasingly favoured for its richer information content and standardised measures. However, extracting quantitative parameters such as the longitudinal relaxation rate (R1), apparent transverse relaxation rate (R2*), or magnetisation-transfer saturation (MTsat) involves inverting a highly non-linear function. Estimations often assume noise-free measurements and use subsets of the data to solve for different quantities in isolation, with error propagating through each computation. Instead, a probabilistic generative model of the entire dataset can be formulated and inverted to jointly recover parameter estimates with a well-defined probabilistic meaning (e.g., maximum likelihood or maximum a posteriori). In practice, iterative methods must be used but convergence is difficult due to the non-convexity of the log-likelihood; yet, we show that it can be achieved thanks to a novel approximate Hessian and, with it, reliable parameter estimates obtained. Here, we demonstrate the utility of this flexible framework in the context of the popular multi-parameter mapping framework and further show how to incorporate a denoising prior and predict posterior uncertainty. Our implementation uses a PyTorch backend and benefits from GPU acceleration. It is available at https://github.com/balbasty/nitorch.
Flexible Bayesian Modelling for Nonlinear Image Registration
Jun 03, 2020



Abstract:We describe a diffeomorphic registration algorithm that allows groups of images to be accurately aligned to a common space, which we intend to incorporate into the SPM software. The idea is to perform inference in a probabilistic graphical model that accounts for variability in both shape and appearance. The resulting framework is general and entirely unsupervised. The model is evaluated at inter-subject registration of 3D human brain scans. Here, the main modeling assumption is that individual anatomies can be generated by deforming a latent 'average' brain. The method is agnostic to imaging modality and can be applied with no prior processing. We evaluate the algorithm using freely available, manually labelled datasets. In this validation we achieve state-of-the-art results, within reasonable runtimes, against previous state-of-the-art widely used, inter-subject registration algorithms. On the unprocessed dataset, the increase in overlap score is over 17%. These results demonstrate the benefits of using informative computational anatomy frameworks for nonlinear registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge