Cathy J. Price
Unified 3D MRI Representations via Sequence-Invariant Contrastive Learning
Jan 21, 2025



Abstract:Self-supervised deep learning has accelerated 2D natural image analysis but remains difficult to translate into 3D MRI, where data are scarce and pre-trained 2D backbones cannot capture volumetric context. We present a sequence-invariant self-supervised framework leveraging quantitative MRI (qMRI). By simulating multiple MRI contrasts from a single 3D qMRI scan and enforcing consistent representations across these contrasts, we learn anatomy-centric rather than sequence-specific features. This yields a robust 3D encoder that performs strongly across varied tasks and protocols. Experiments on healthy brain segmentation (IXI), stroke lesion segmentation (ARC), and MRI denoising show significant gains over baseline SSL approaches, especially in low-data settings (up to +8.3% Dice, +4.2 dB PSNR). Our model also generalises effectively to unseen sites, demonstrating potential for more scalable and clinically reliable volumetric analysis. All code and trained models are publicly available.
Domain-Agnostic Stroke Lesion Segmentation Using Physics-Constrained Synthetic Data
Dec 04, 2024



Abstract:Segmenting stroke lesions in Magnetic Resonance Imaging (MRI) is challenging due to diverse clinical imaging domains, with existing models struggling to generalise across different MRI acquisition parameters and sequences. In this work, we propose two novel physics-constrained approaches using synthetic quantitative MRI (qMRI) images to enhance the robustness and generalisability of segmentation models. We trained a qMRI estimation model to predict qMRI maps from MPRAGE images, which were used to simulate diverse MRI sequences for segmentation training. A second approach built upon prior work in synthetic data for stroke lesion segmentation, generating qMRI maps from a dataset of tissue labels. The proposed approaches improved over the baseline nnUNet on a variety of out-of-distribution datasets, with the second approach outperforming the prior synthetic data method.
Synthetic Data for Robust Stroke Segmentation
Apr 02, 2024Abstract:Deep learning-based semantic segmentation in neuroimaging currently requires high-resolution scans and extensive annotated datasets, posing significant barriers to clinical applicability. We present a novel synthetic framework for the task of lesion segmentation, extending the capabilities of the established SynthSeg approach to accommodate large heterogeneous pathologies with lesion-specific augmentation strategies. Our method trains deep learning models, demonstrated here with the UNet architecture, using label maps derived from healthy and stroke datasets, facilitating the segmentation of both healthy tissue and pathological lesions without sequence-specific training data. Evaluated against in-domain and out-of-domain (OOD) datasets, our framework demonstrates robust performance, rivaling current methods within the training domain and significantly outperforming them on OOD data. This contribution holds promise for advancing medical imaging analysis in clinical settings, especially for stroke pathology, by enabling reliable segmentation across varied imaging sequences with reduced dependency on large annotated corpora. Code and weights available at https://github.com/liamchalcroft/SynthStroke.
Predicting recovery following stroke: deep learning, multimodal data and feature selection using explainable AI
Oct 29, 2023



Abstract:Machine learning offers great potential for automated prediction of post-stroke symptoms and their response to rehabilitation. Major challenges for this endeavour include the very high dimensionality of neuroimaging data, the relatively small size of the datasets available for learning, and how to effectively combine neuroimaging and tabular data (e.g. demographic information and clinical characteristics). This paper evaluates several solutions based on two strategies. The first is to use 2D images that summarise MRI scans. The second is to select key features that improve classification accuracy. Additionally, we introduce the novel approach of training a convolutional neural network (CNN) on images that combine regions-of-interest extracted from MRIs, with symbolic representations of tabular data. We evaluate a series of CNN architectures (both 2D and a 3D) that are trained on different representations of MRI and tabular data, to predict whether a composite measure of post-stroke spoken picture description ability is in the aphasic or non-aphasic range. MRI and tabular data were acquired from 758 English speaking stroke survivors who participated in the PLORAS study. The classification accuracy for a baseline logistic regression was 0.678 for lesion size alone, rising to 0.757 and 0.813 when initial symptom severity and recovery time were successively added. The highest classification accuracy 0.854 was observed when 8 regions-of-interest was extracted from each MRI scan and combined with lesion size, initial severity and recovery time in a 2D Residual Neural Network.Our findings demonstrate how imaging and tabular data can be combined for high post-stroke classification accuracy, even when the dataset is small in machine learning terms. We conclude by proposing how the current models could be improved to achieve even higher levels of accuracy using images from hospital scanners.
Large-kernel Attention for Efficient and Robust Brain Lesion Segmentation
Aug 14, 2023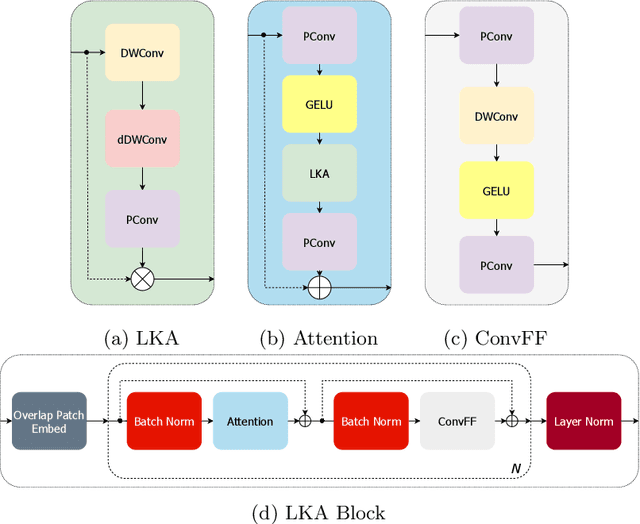
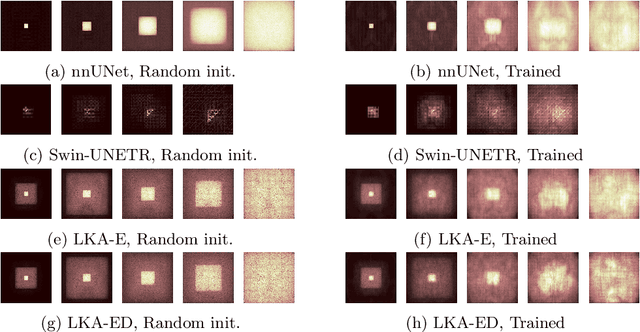
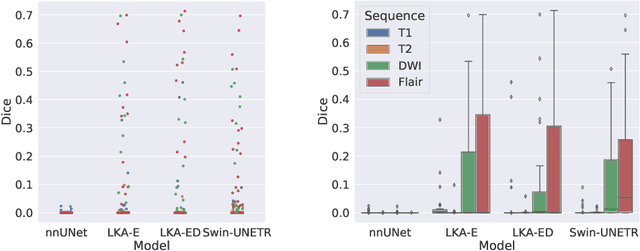
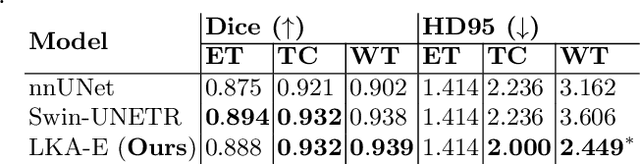
Abstract:Vision transformers are effective deep learning models for vision tasks, including medical image segmentation. However, they lack efficiency and translational invariance, unlike convolutional neural networks (CNNs). To model long-range interactions in 3D brain lesion segmentation, we propose an all-convolutional transformer block variant of the U-Net architecture. We demonstrate that our model provides the greatest compromise in three factors: performance competitive with the state-of-the-art; parameter efficiency of a CNN; and the favourable inductive biases of a transformer. Our public implementation is available at https://github.com/liamchalcroft/MDUNet .
Predicting Language Recovery after Stroke with Convolutional Networks on Stitched MRI
Nov 26, 2018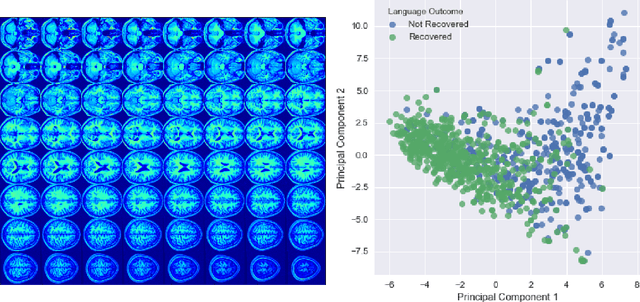

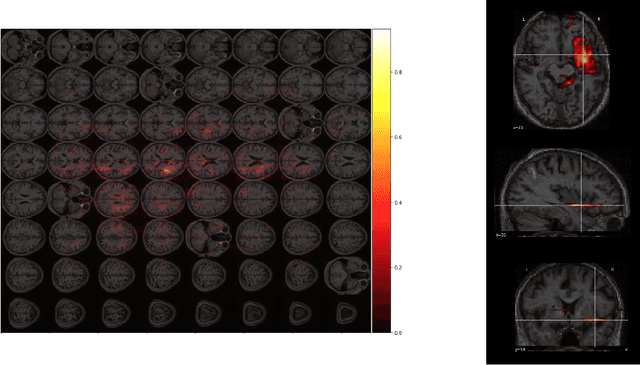
Abstract:One third of stroke survivors have language difficulties. Emerging evidence suggests that their likelihood of recovery depends mainly on the damage to language centers. Thus previous research for predicting language recovery post-stroke has focused on identifying damaged regions of the brain. In this paper, we introduce a novel method where we only make use of stitched 2-dimensional cross-sections of raw MRI scans in a deep convolutional neural network setup to predict language recovery post-stroke. Our results show: a) the proposed model that only uses MRI scans has comparable performance to models that are dependent on lesion specific information; b) the features learned by our model are complementary to the lesion specific information and the combination of both appear to outperform previously reported results in similar settings. We further analyse the CNN model for understanding regions in brain that are responsible for arriving at these predictions using gradient based saliency maps. Our findings are in line with previous lesion studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge