Liam Chalcroft
Unified 3D MRI Representations via Sequence-Invariant Contrastive Learning
Jan 21, 2025



Abstract:Self-supervised deep learning has accelerated 2D natural image analysis but remains difficult to translate into 3D MRI, where data are scarce and pre-trained 2D backbones cannot capture volumetric context. We present a sequence-invariant self-supervised framework leveraging quantitative MRI (qMRI). By simulating multiple MRI contrasts from a single 3D qMRI scan and enforcing consistent representations across these contrasts, we learn anatomy-centric rather than sequence-specific features. This yields a robust 3D encoder that performs strongly across varied tasks and protocols. Experiments on healthy brain segmentation (IXI), stroke lesion segmentation (ARC), and MRI denoising show significant gains over baseline SSL approaches, especially in low-data settings (up to +8.3% Dice, +4.2 dB PSNR). Our model also generalises effectively to unseen sites, demonstrating potential for more scalable and clinically reliable volumetric analysis. All code and trained models are publicly available.
Domain-Agnostic Stroke Lesion Segmentation Using Physics-Constrained Synthetic Data
Dec 04, 2024



Abstract:Segmenting stroke lesions in Magnetic Resonance Imaging (MRI) is challenging due to diverse clinical imaging domains, with existing models struggling to generalise across different MRI acquisition parameters and sequences. In this work, we propose two novel physics-constrained approaches using synthetic quantitative MRI (qMRI) images to enhance the robustness and generalisability of segmentation models. We trained a qMRI estimation model to predict qMRI maps from MPRAGE images, which were used to simulate diverse MRI sequences for segmentation training. A second approach built upon prior work in synthetic data for stroke lesion segmentation, generating qMRI maps from a dataset of tissue labels. The proposed approaches improved over the baseline nnUNet on a variety of out-of-distribution datasets, with the second approach outperforming the prior synthetic data method.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024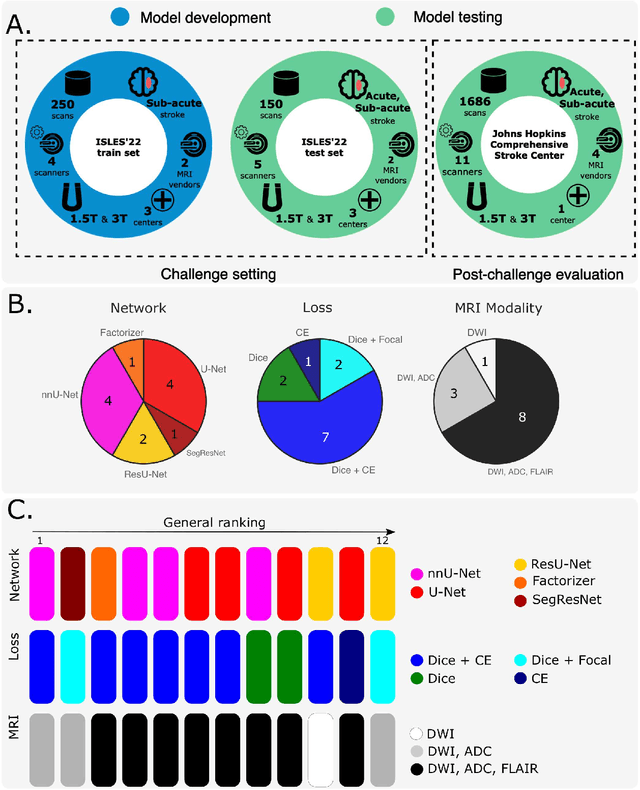
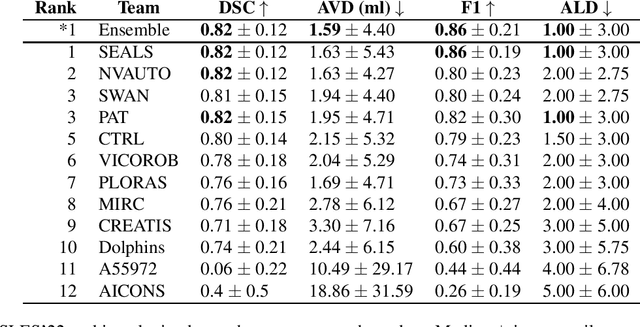
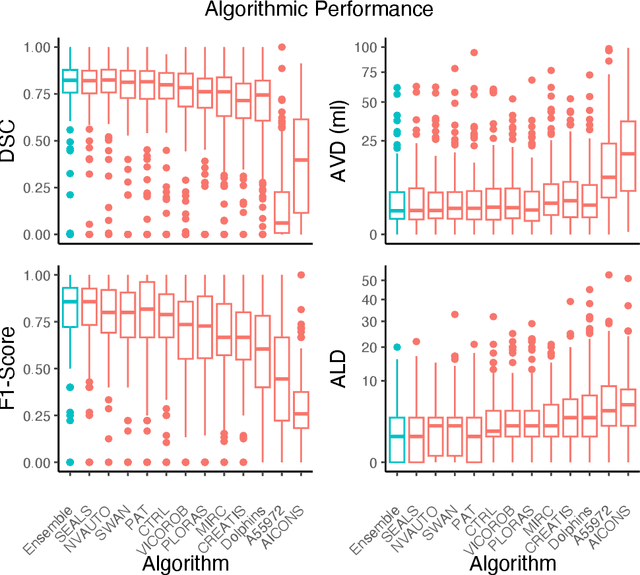
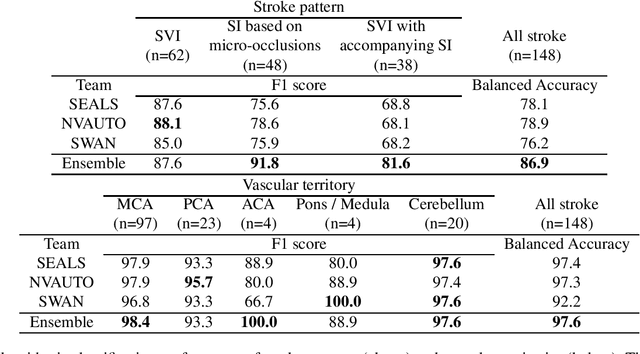
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Synthetic Data for Robust Stroke Segmentation
Apr 02, 2024Abstract:Deep learning-based semantic segmentation in neuroimaging currently requires high-resolution scans and extensive annotated datasets, posing significant barriers to clinical applicability. We present a novel synthetic framework for the task of lesion segmentation, extending the capabilities of the established SynthSeg approach to accommodate large heterogeneous pathologies with lesion-specific augmentation strategies. Our method trains deep learning models, demonstrated here with the UNet architecture, using label maps derived from healthy and stroke datasets, facilitating the segmentation of both healthy tissue and pathological lesions without sequence-specific training data. Evaluated against in-domain and out-of-domain (OOD) datasets, our framework demonstrates robust performance, rivaling current methods within the training domain and significantly outperforming them on OOD data. This contribution holds promise for advancing medical imaging analysis in clinical settings, especially for stroke pathology, by enabling reliable segmentation across varied imaging sequences with reduced dependency on large annotated corpora. Code and weights available at https://github.com/liamchalcroft/SynthStroke.
Large-kernel Attention for Efficient and Robust Brain Lesion Segmentation
Aug 14, 2023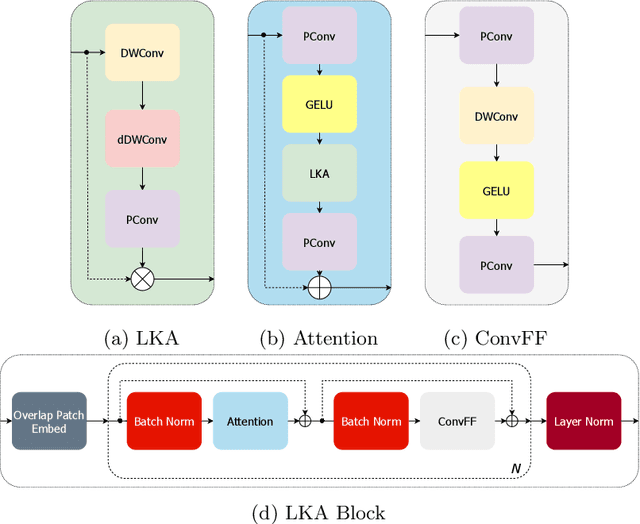
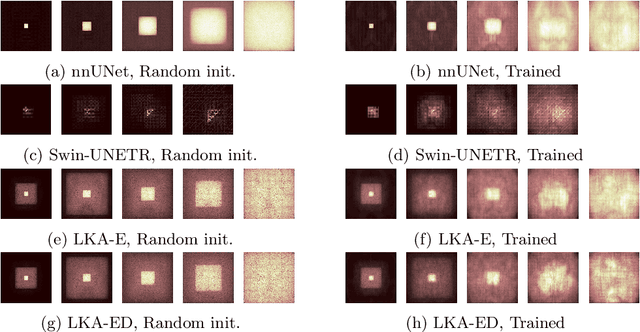
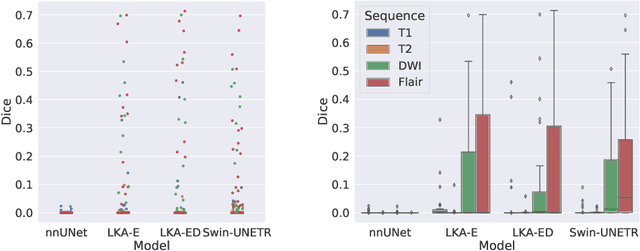
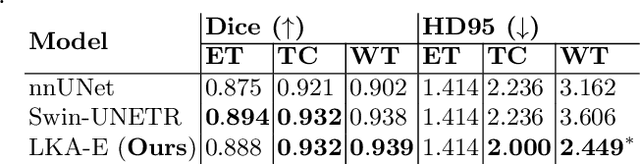
Abstract:Vision transformers are effective deep learning models for vision tasks, including medical image segmentation. However, they lack efficiency and translational invariance, unlike convolutional neural networks (CNNs). To model long-range interactions in 3D brain lesion segmentation, we propose an all-convolutional transformer block variant of the U-Net architecture. We demonstrate that our model provides the greatest compromise in three factors: performance competitive with the state-of-the-art; parameter efficiency of a CNN; and the favourable inductive biases of a transformer. Our public implementation is available at https://github.com/liamchalcroft/MDUNet .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge