Geraint Rees
Generalizable automated ischaemic stroke lesion segmentation with vision transformers
Feb 10, 2025
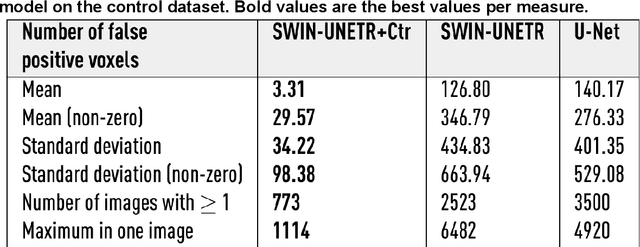
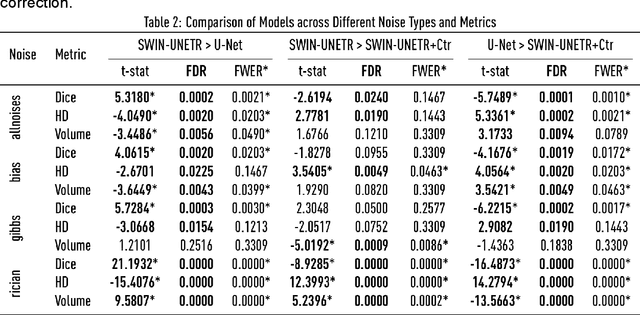
Abstract:Ischaemic stroke, a leading cause of death and disability, critically relies on neuroimaging for characterising the anatomical pattern of injury. Diffusion-weighted imaging (DWI) provides the highest expressivity in ischemic stroke but poses substantial challenges for automated lesion segmentation: susceptibility artefacts, morphological heterogeneity, age-related comorbidities, time-dependent signal dynamics, instrumental variability, and limited labelled data. Current U-Net-based models therefore underperform, a problem accentuated by inadequate evaluation metrics that focus on mean performance, neglecting anatomical, subpopulation, and acquisition-dependent variability. Here, we present a high-performance DWI lesion segmentation tool addressing these challenges through optimized vision transformer-based architectures, integration of 3563 annotated lesions from multi-site data, and algorithmic enhancements, achieving state-of-the-art results. We further propose a novel evaluative framework assessing model fidelity, equity (across demographics and lesion subtypes), anatomical precision, and robustness to instrumental variability, promoting clinical and research utility. This work advances stroke imaging by reconciling model expressivity with domain-specific challenges and redefining performance benchmarks to prioritize equity and generalizability, critical for personalized medicine and mechanistic research.
The legibility of the imaged human brain
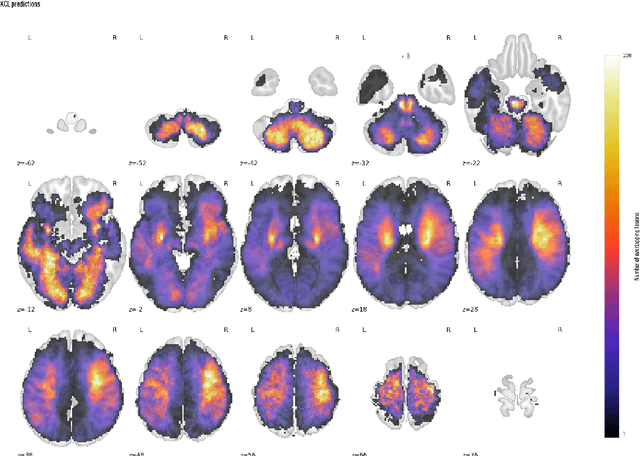
Aug 23, 2023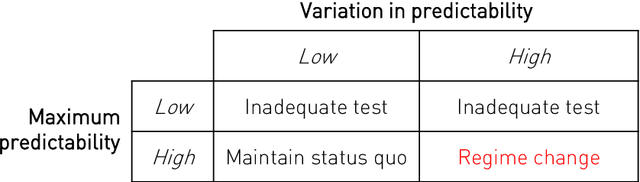

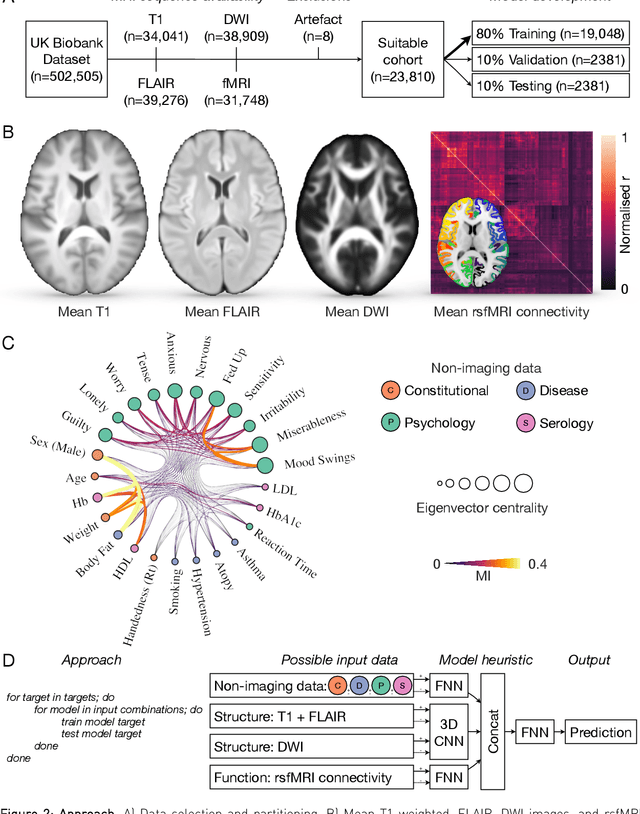
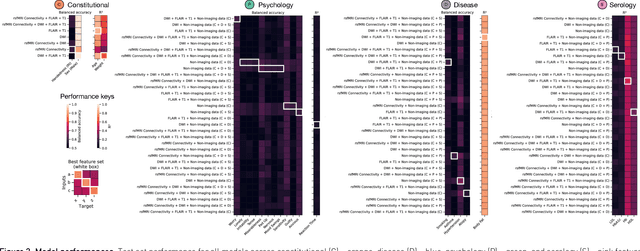
Abstract:Our knowledge of the organisation of the human brain at the population-level is yet to translate into power to predict functional differences at the individual-level, limiting clinical applications, and casting doubt on the generalisability of inferred mechanisms. It remains unknown whether the difficulty arises from the absence of individuating biological patterns within the brain, or from limited power to access them with the models and compute at our disposal. Here we comprehensively investigate the resolvability of such patterns with data and compute at unprecedented scale. Across 23810 unique participants from UK Biobank, we systematically evaluate the predictability of 25 individual biological characteristics, from all available combinations of structural and functional neuroimaging data. Over 4526 GPU*hours of computation, we train, optimize, and evaluate out-of-sample 700 individual predictive models, including multilayer perceptrons of demographic, psychological, serological, chronic morbidity, and functional connectivity characteristics, and both uni- and multi-modal 3D convolutional neural network models of macro- and micro-structural brain imaging. We find a marked discrepancy between the high predictability of sex (balanced accuracy 99.7%), age (mean absolute error 2.048 years, R2 0.859), and weight (mean absolute error 2.609Kg, R2 0.625), for which we set new state-of-the-art performance, and the surprisingly low predictability of other characteristics. Neither structural nor functional imaging predicted individual psychology better than the coincidence of common chronic morbidity (p<0.05). Serology predicted common morbidity (p<0.05) and was best predicted by it (p<0.001), followed by structural neuroimaging (p<0.05). Our findings suggest either more informative imaging or more powerful models will be needed to decipher individual level characteristics from the brain.
Representational Ethical Model Calibration
Jul 25, 2022



Abstract:Equity is widely held to be fundamental to the ethics of healthcare. In the context of clinical decision-making, it rests on the comparative fidelity of the intelligence -- evidence-based or intuitive -- guiding the management of each individual patient. Though brought to recent attention by the individuating power of contemporary machine learning, such epistemic equity arises in the context of any decision guidance, whether traditional or innovative. Yet no general framework for its quantification, let alone assurance, currently exists. Here we formulate epistemic equity in terms of model fidelity evaluated over learnt multi-dimensional representations of identity crafted to maximise the captured diversity of the population, introducing a comprehensive framework for Representational Ethical Model Calibration. We demonstrate use of the framework on large-scale multimodal data from UK Biobank to derive diverse representations of the population, quantify model performance, and institute responsive remediation. We offer our approach as a principled solution to quantifying and assuring epistemic equity in healthcare, with applications across the research, clinical, and regulatory domains.
Fitting Segmentation Networks on Varying Image Resolutions using Splatting
Jun 15, 2022


Abstract:Data used in image segmentation are not always defined on the same grid. This is particularly true for medical images, where the resolution, field-of-view and orientation can differ across channels and subjects. Images and labels are therefore commonly resampled onto the same grid, as a pre-processing step. However, the resampling operation introduces partial volume effects and blurring, thereby changing the effective resolution and reducing the contrast between structures. In this paper we propose a splat layer, which automatically handles resolution mismatches in the input data. This layer pushes each image onto a mean space where the forward pass is performed. As the splat operator is the adjoint to the resampling operator, the mean-space prediction can be pulled back to the native label space, where the loss function is computed. Thus, the need for explicit resolution adjustment using interpolation is removed. We show on two publicly available datasets, with simulated and real multi-modal magnetic resonance images, that this model improves segmentation results compared to resampling as a pre-processing step.
Fast Unsupervised Brain Anomaly Detection and Segmentation with Diffusion Models
Jun 07, 2022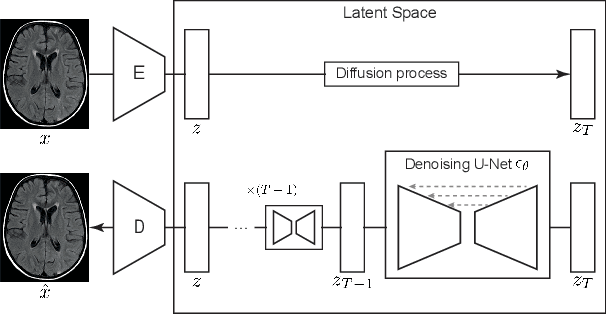
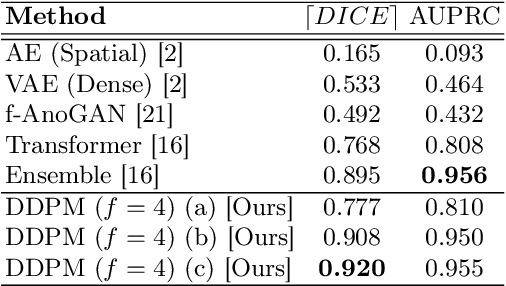
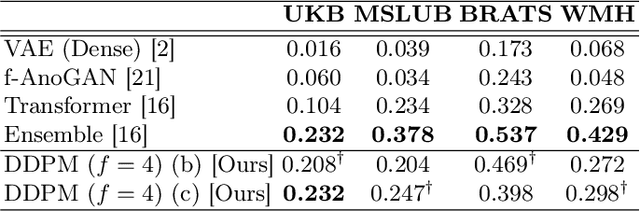
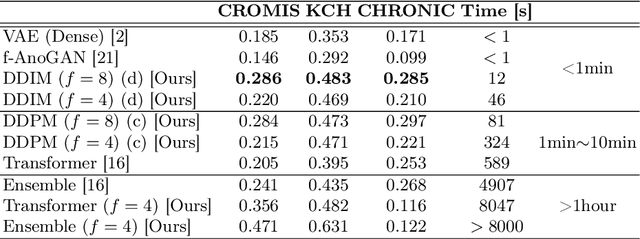
Abstract:Deep generative models have emerged as promising tools for detecting arbitrary anomalies in data, dispensing with the necessity for manual labelling. Recently, autoregressive transformers have achieved state-of-the-art performance for anomaly detection in medical imaging. Nonetheless, these models still have some intrinsic weaknesses, such as requiring images to be modelled as 1D sequences, the accumulation of errors during the sampling process, and the significant inference times associated with transformers. Denoising diffusion probabilistic models are a class of non-autoregressive generative models recently shown to produce excellent samples in computer vision (surpassing Generative Adversarial Networks), and to achieve log-likelihoods that are competitive with transformers while having fast inference times. Diffusion models can be applied to the latent representations learnt by autoencoders, making them easily scalable and great candidates for application to high dimensional data, such as medical images. Here, we propose a method based on diffusion models to detect and segment anomalies in brain imaging. By training the models on healthy data and then exploring its diffusion and reverse steps across its Markov chain, we can identify anomalous areas in the latent space and hence identify anomalies in the pixel space. Our diffusion models achieve competitive performance compared with autoregressive approaches across a series of experiments with 2D CT and MRI data involving synthetic and real pathological lesions with much reduced inference times, making their usage clinically viable.
Equitable modelling of brain imaging by counterfactual augmentation with morphologically constrained 3D deep generative models
Nov 29, 2021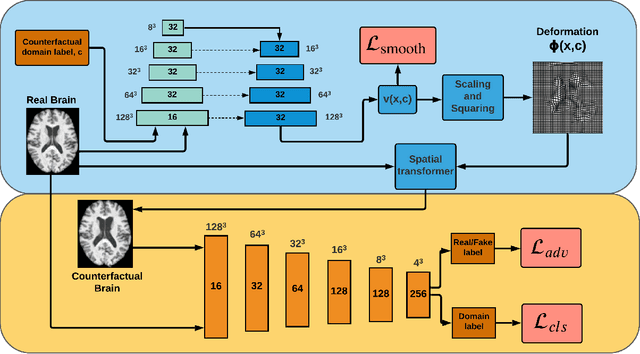
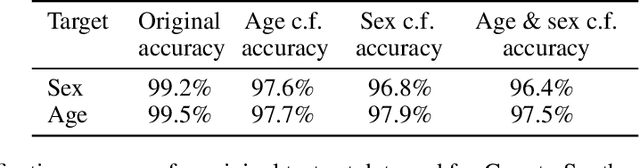
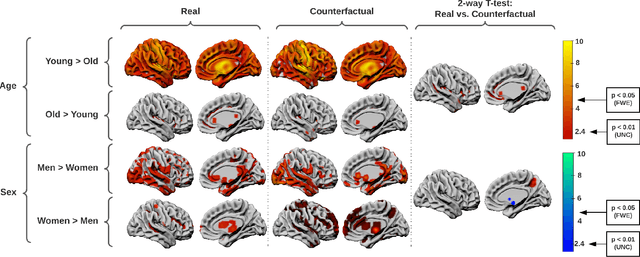
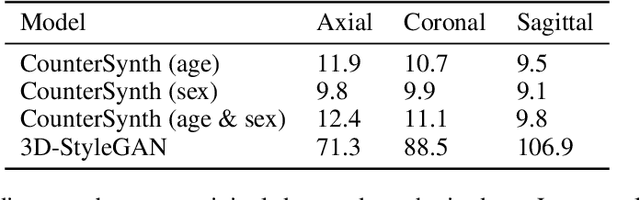
Abstract:We describe Countersynth, a conditional generative model of diffeomorphic deformations that induce label-driven, biologically plausible changes in volumetric brain images. The model is intended to synthesise counterfactual training data augmentations for downstream discriminative modelling tasks where fidelity is limited by data imbalance, distributional instability, confounding, or underspecification, and exhibits inequitable performance across distinct subpopulations. Focusing on demographic attributes, we evaluate the quality of synthesized counterfactuals with voxel-based morphometry, classification and regression of the conditioning attributes, and the Fr\'{e}chet inception distance. Examining downstream discriminative performance in the context of engineered demographic imbalance and confounding, we use UK Biobank magnetic resonance imaging data to benchmark CounterSynth augmentation against current solutions to these problems. We achieve state-of-the-art improvements, both in overall fidelity and equity. The source code for CounterSynth is available online.
An MRF-UNet Product of Experts for Image Segmentation
Apr 12, 2021



Abstract:While convolutional neural networks (CNNs) trained by back-propagation have seen unprecedented success at semantic segmentation tasks, they are known to struggle on out-of-distribution data. Markov random fields (MRFs) on the other hand, encode simpler distributions over labels that, although less flexible than UNets, are less prone to over-fitting. In this paper, we propose to fuse both strategies by computing the product of distributions of a UNet and an MRF. As this product is intractable, we solve for an approximate distribution using an iterative mean-field approach. The resulting MRF-UNet is trained jointly by back-propagation. Compared to other works using conditional random fields (CRFs), the MRF has no dependency on the imaging data, which should allow for less over-fitting. We show on 3D neuroimaging data that this novel network improves generalisation to out-of-distribution samples. Furthermore, it allows the overall number of parameters to be reduced while preserving high accuracy. These results suggest that a classic MRF smoothness prior can allow for less over-fitting when principally integrated into a CNN model. Our implementation is available at https://github.com/balbasty/nitorch.
Unsupervised Brain Anomaly Detection and Segmentation with Transformers
Feb 23, 2021



Abstract:Pathological brain appearances may be so heterogeneous as to be intelligible only as anomalies, defined by their deviation from normality rather than any specific pathological characteristic. Amongst the hardest tasks in medical imaging, detecting such anomalies requires models of the normal brain that combine compactness with the expressivity of the complex, long-range interactions that characterise its structural organisation. These are requirements transformers have arguably greater potential to satisfy than other current candidate architectures, but their application has been inhibited by their demands on data and computational resource. Here we combine the latent representation of vector quantised variational autoencoders with an ensemble of autoregressive transformers to enable unsupervised anomaly detection and segmentation defined by deviation from healthy brain imaging data, achievable at low computational cost, within relative modest data regimes. We compare our method to current state-of-the-art approaches across a series of experiments involving synthetic and real pathological lesions. On real lesions, we train our models on 15,000 radiologically normal participants from UK Biobank, and evaluate performance on four different brain MR datasets with small vessel disease, demyelinating lesions, and tumours. We demonstrate superior anomaly detection performance both image-wise and pixel-wise, achievable without post-processing. These results draw attention to the potential of transformers in this most challenging of imaging tasks.
Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy
Sep 12, 2018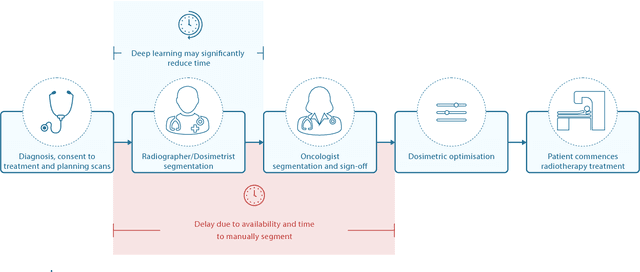
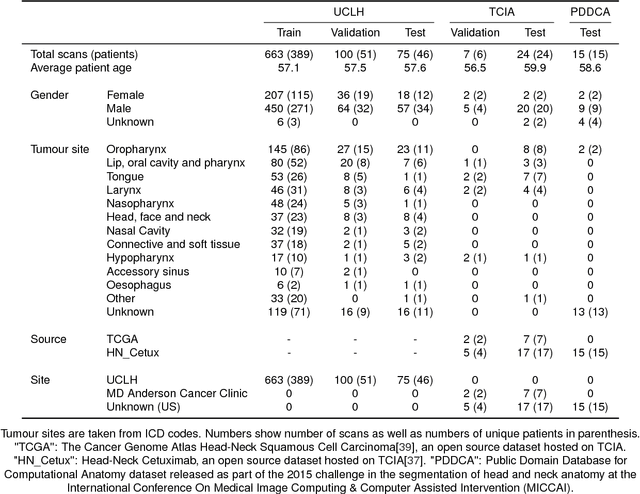
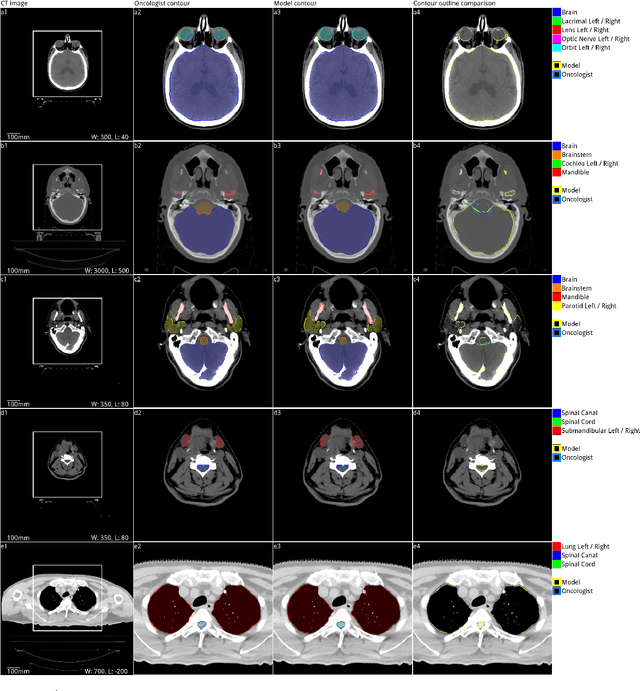
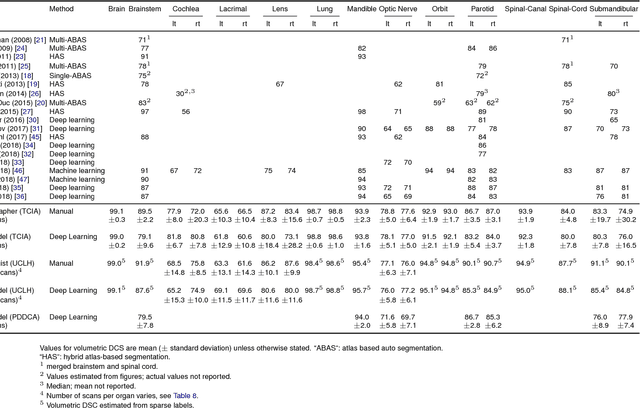
Abstract:Over half a million individuals are diagnosed with head and neck cancer each year worldwide. Radiotherapy is an important curative treatment for this disease, but it requires manually intensive delineation of radiosensitive organs at risk (OARs). This planning process can delay treatment commencement. While auto-segmentation algorithms offer a potentially time-saving solution, the challenges in defining, quantifying and achieving expert performance remain. Adopting a deep learning approach, we demonstrate a 3D U-Net architecture that achieves performance similar to experts in delineating a wide range of head and neck OARs. The model was trained on a dataset of 663 deidentified computed tomography (CT) scans acquired in routine clinical practice and segmented according to consensus OAR definitions. We demonstrate its generalisability through application to an independent test set of 24 CT scans available from The Cancer Imaging Archive collected at multiple international sites previously unseen to the model, each segmented by two independent experts and consisting of 21 OARs commonly segmented in clinical practice. With appropriate validation studies and regulatory approvals, this system could improve the effectiveness of radiotherapy pathways.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge